Reduce Operators with Pangeo#
Reduce Operators#
When computing statistics over time or indices based on multiple bands, it is possible to use reduce operators.
In Pangeo and Xarray we can use the groupby method, which applies a reducer to a data cube dimension by collapsing all the values along the specified dimension into an output value computed by the reducer.
Reduce the temporal dimension to a single value, the mean for instance:
# STAC Catalogue Libraries
import pystac_client
import stackstac
import xarray as xr
# West, South, East, North
spatial_extent = [11.259613, 46.461019, 11.406212, 46.522237]
temporal_extent = ["2021-05-28T00:00:00Z","2021-06-30T00:00:00Z"]
URL = "https://earth-search.aws.element84.com/v1"
catalog = pystac_client.Client.open(URL)
s2_items = catalog.search(
bbox=spatial_extent,
datetime=temporal_extent,
collections=["sentinel-2-l2a"]
).item_collection()
s2_cube = stackstac.stack(s2_items,
bounds_latlon=spatial_extent,
assets=["red","nir"]
)
s2_cube
/srv/conda/envs/notebook/lib/python3.11/site-packages/stackstac/prepare.py:408: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
<xarray.DataArray 'stackstac-b671372a6175b16d2e4eaf48f5606039' (time: 26,
band: 2,
y: 714, x: 1146)>
dask.array<fetch_raster_window, shape=(26, 2, 714, 1146), dtype=float64, chunksize=(1, 1, 714, 1024), chunktype=numpy.ndarray>
Coordinates: (12/53)
* time (time) datetime64[ns] 2021-05-28...
id (time) <U24 'S2B_32TPS_20210528_...
* band (band) <U3 'red' 'nir'
* x (x) float64 6.733e+05 ... 6.848e+05
* y (y) float64 5.155e+06 ... 5.148e+06
s2:unclassified_percentage (time) float64 0.2507 ... 2.393
... ...
proj:transform object {0, 600000, 10, 5200020, ...
title (band) <U20 'Red (band 4) - 10m'...
common_name (band) <U3 'red' 'nir'
center_wavelength (band) float64 0.665 0.842
full_width_half_max (band) float64 0.038 0.145
epsg int64 32632
Attributes:
spec: RasterSpec(epsg=32632, bounds=(673310.0, 5147750.0, 684770.0...
crs: epsg:32632
transform: | 10.00, 0.00, 673310.00|\n| 0.00,-10.00, 5154890.00|\n| 0.0...
resolution: 10.0- time: 26
- band: 2
- y: 714
- x: 1146
- dask.array<chunksize=(1, 1, 714, 1024), meta=np.ndarray>
Array Chunk Bytes 324.62 MiB 5.58 MiB Shape (26, 2, 714, 1146) (1, 1, 714, 1024) Dask graph 104 chunks in 3 graph layers Data type float64 numpy.ndarray - time(time)datetime64[ns]2021-05-28T10:17:59.882000 ... 2...
array(['2021-05-28T10:17:59.882000000', '2021-05-28T10:17:59.884000000', '2021-05-31T10:27:56.286000000', '2021-05-31T10:27:56.289000000', '2021-06-02T10:18:00.103000000', '2021-06-02T10:18:00.104000000', '2021-06-05T10:27:56.280000000', '2021-06-05T10:27:56.282000000', '2021-06-07T10:18:00.323000000', '2021-06-07T10:18:00.323000000', '2021-06-10T10:27:56.565000000', '2021-06-10T10:27:56.566000000', '2021-06-12T10:17:59.823000000', '2021-06-12T10:17:59.824000000', '2021-06-15T10:27:55.829000000', '2021-06-15T10:27:55.830000000', '2021-06-17T10:18:00.191000000', '2021-06-17T10:18:00.192000000', '2021-06-20T10:27:56.235000000', '2021-06-20T10:27:56.236000000', '2021-06-22T10:18:00.204000000', '2021-06-22T10:18:00.205000000', '2021-06-25T10:27:56.865000000', '2021-06-25T10:27:56.867000000', '2021-06-27T10:18:00.077000000', '2021-06-27T10:18:00.078000000'], dtype='datetime64[ns]') - id(time)<U24'S2B_32TPS_20210528_1_L2A' ... '...
array(['S2B_32TPS_20210528_1_L2A', 'S2B_32TPS_20210528_0_L2A', 'S2B_32TPS_20210531_1_L2A', 'S2B_32TPS_20210531_0_L2A', 'S2A_32TPS_20210602_1_L2A', 'S2A_32TPS_20210602_0_L2A', 'S2A_32TPS_20210605_0_L2A', 'S2A_32TPS_20210605_1_L2A', 'S2B_32TPS_20210607_1_L2A', 'S2B_32TPS_20210607_0_L2A', 'S2B_32TPS_20210610_1_L2A', 'S2B_32TPS_20210610_0_L2A', 'S2A_32TPS_20210612_1_L2A', 'S2A_32TPS_20210612_0_L2A', 'S2A_32TPS_20210615_0_L2A', 'S2A_32TPS_20210615_1_L2A', 'S2B_32TPS_20210617_1_L2A', 'S2B_32TPS_20210617_0_L2A', 'S2B_32TPS_20210620_0_L2A', 'S2B_32TPS_20210620_1_L2A', 'S2A_32TPS_20210622_1_L2A', 'S2A_32TPS_20210622_0_L2A', 'S2A_32TPS_20210625_0_L2A', 'S2A_32TPS_20210625_1_L2A', 'S2B_32TPS_20210627_1_L2A', 'S2B_32TPS_20210627_0_L2A'], dtype='<U24') - band(band)<U3'red' 'nir'
array(['red', 'nir'], dtype='<U3')
- x(x)float646.733e+05 6.733e+05 ... 6.848e+05
array([673310., 673320., 673330., ..., 684740., 684750., 684760.])
- y(y)float645.155e+06 5.155e+06 ... 5.148e+06
array([5154890., 5154880., 5154870., ..., 5147780., 5147770., 5147760.])
- s2:unclassified_percentage(time)float640.2507 6.344 ... 0.3769 2.393
array([0.250706, 6.3441 , 0.515761, 3.860853, 1.493286, 5.219495, 4.114496, 1.357601, 0.809151, 1.985232, 2.379116, 4.723761, 2.041206, 4.504375, 2.619704, 0.152735, 0.65733 , 3.93955 , 0.770469, 0.292153, 0.427856, 3.074201, 3.535479, 0.935654, 0.376936, 2.392862]) - s2:product_type()<U7'S2MSI2A'
array('S2MSI2A', dtype='<U7') - s2:sequence(time)<U1'1' '0' '1' '0' ... '0' '1' '1' '0'
array(['1', '0', '1', '0', '1', '0', '0', '1', '1', '0', '1', '0', '1', '0', '0', '1', '1', '0', '0', '1', '1', '0', '0', '1', '1', '0'], dtype='<U1') - s2:generation_time(time)<U27'2023-02-27T08:23:12.000000Z' .....
array(['2023-02-27T08:23:12.000000Z', '2021-05-28T13:08:36.000000Z', '2023-03-01T00:24:41.000000Z', '2021-05-31T14:00:40.000000Z', '2023-03-13T04:04:11.000000Z', '2021-06-02T17:13:07.000000Z', '2021-06-05T15:23:29.000000Z', '2023-03-12T16:27:26.000000Z', '2023-03-14T07:14:12.000000Z', '2021-06-07T16:37:34.000000Z', '2023-03-21T10:24:57.000000Z', '2021-06-10T13:27:39.000000Z', '2023-03-16T11:37:02.000000Z', '2021-06-12T13:18:42.000000Z', '2021-06-15T13:16:59.000000Z', '2023-03-21T22:10:50.000000Z', '2023-03-21T22:17:28.000000Z', '2021-06-17T13:12:19.000000Z', '2021-06-20T14:05:52.000000Z', '2023-03-08T17:05:00.000000Z', '2023-01-29T15:30:31.000000Z', '2021-06-22T12:13:05.000000Z', '2021-06-25T13:25:02.000000Z', '2023-03-19T05:49:11.000000Z', '2023-03-18T23:38:59.000000Z', '2021-06-27T13:58:56.000000Z'], dtype='<U27') - s2:datatake_type()<U8'INS-NOBS'
array('INS-NOBS', dtype='<U8') - proj:epsg()int6432632
array(32632)
- mgrs:latitude_band()<U1'T'
array('T', dtype='<U1') - processing:software(time)object{'sentinel2-to-stac': '0.1.1'} ....
array([{'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}], dtype=object) - s2:processing_baseline(time)<U5'05.00' '03.00' ... '05.00' '03.00'
array(['05.00', '03.00', '05.00', '03.00', '05.00', '03.00', '03.00', '05.00', '05.00', '03.00', '05.00', '03.00', '05.00', '03.00', '03.00', '05.00', '05.00', '03.00', '03.00', '05.00', '05.00', '03.00', '03.00', '05.00', '05.00', '03.00'], dtype='<U5') - s2:datastrip_id(time)<U64'S2B_OPER_MSI_L2A_DS_S2RP_202302...
array(['S2B_OPER_MSI_L2A_DS_S2RP_20230227T082312_S20210528T100555_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20210528T130836_S20210528T100555_N03.00', 'S2B_OPER_MSI_L2A_DS_S2RP_20230301T002441_S20210531T102618_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS4_20210531T140040_S20210531T102618_N03.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20230313T040411_S20210602T101235_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210602T171307_S20210602T101235_N03.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210605T152329_S20210605T102737_N03.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20230312T162726_S20210605T102737_N05.00', 'S2B_OPER_MSI_L2A_DS_S2RP_20230314T071412_S20210607T101225_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20210607T163734_S20210607T101225_N03.00', 'S2B_OPER_MSI_L2A_DS_S2RP_20230321T102457_S20210610T102220_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20210610T132739_S20210610T102220_N03.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20230316T113702_S20210612T101329_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210612T131842_S20210612T101329_N03.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210615T131659_S20210615T102602_N03.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20230321T221050_S20210615T102602_N05.00', 'S2B_OPER_MSI_L2A_DS_S2RP_20230321T221728_S20210617T101301_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20210617T131219_S20210617T101301_N03.00', 'S2B_OPER_MSI_L2A_DS_VGS4_20210620T140552_S20210620T102722_N03.00', 'S2B_OPER_MSI_L2A_DS_S2RP_20230308T170500_S20210620T102722_N05.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20230129T153031_S20210622T101416_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS4_20210622T121305_S20210622T101416_N03.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210625T132502_S20210625T102545_N03.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20230319T054911_S20210625T102545_N05.00', 'S2B_OPER_MSI_L2A_DS_S2RP_20230318T233859_S20210627T101702_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS4_20210627T135856_S20210627T101702_N03.00'], dtype='<U64') - s2:product_uri(time)<U65'S2B_MSIL2A_20210528T100559_N050...
array(['S2B_MSIL2A_20210528T100559_N0500_R022_T32TPS_20230227T082312.SAFE', 'S2B_MSIL2A_20210528T100559_N0300_R022_T32TPS_20210528T130836.SAFE', 'S2B_MSIL2A_20210531T101559_N0500_R065_T32TPS_20230301T002441.SAFE', 'S2B_MSIL2A_20210531T101559_N0300_R065_T32TPS_20210531T140040.SAFE', 'S2A_MSIL2A_20210602T101031_N0500_R022_T32TPS_20230313T040411.SAFE', 'S2A_MSIL2A_20210602T101031_N0300_R022_T32TPS_20210602T171307.SAFE', 'S2A_MSIL2A_20210605T102021_N0300_R065_T32TPS_20210605T152329.SAFE', 'S2A_MSIL2A_20210605T102021_N0500_R065_T32TPS_20230312T162726.SAFE', 'S2B_MSIL2A_20210607T100559_N0500_R022_T32TPS_20230314T071412.SAFE', 'S2B_MSIL2A_20210607T100559_N0300_R022_T32TPS_20210607T163734.SAFE', 'S2B_MSIL2A_20210610T101559_N0500_R065_T32TPS_20230321T102457.SAFE', 'S2B_MSIL2A_20210610T101559_N0300_R065_T32TPS_20210610T132739.SAFE', 'S2A_MSIL2A_20210612T101031_N0500_R022_T32TPS_20230316T113702.SAFE', 'S2A_MSIL2A_20210612T101031_N0300_R022_T32TPS_20210612T131842.SAFE', 'S2A_MSIL2A_20210615T102021_N0300_R065_T32TPS_20210615T131659.SAFE', 'S2A_MSIL2A_20210615T102021_N0500_R065_T32TPS_20230321T221050.SAFE', 'S2B_MSIL2A_20210617T100559_N0500_R022_T32TPS_20230321T221728.SAFE', 'S2B_MSIL2A_20210617T100559_N0300_R022_T32TPS_20210617T131219.SAFE', 'S2B_MSIL2A_20210620T101559_N0300_R065_T32TPS_20210620T140552.SAFE', 'S2B_MSIL2A_20210620T101559_N0500_R065_T32TPS_20230308T170500.SAFE', 'S2A_MSIL2A_20210622T101031_N0500_R022_T32TPS_20230129T153031.SAFE', 'S2A_MSIL2A_20210622T101031_N0300_R022_T32TPS_20210622T121305.SAFE', 'S2A_MSIL2A_20210625T102021_N0300_R065_T32TPS_20210625T132502.SAFE', 'S2A_MSIL2A_20210625T102021_N0500_R065_T32TPS_20230319T054911.SAFE', 'S2B_MSIL2A_20210627T100559_N0500_R022_T32TPS_20230318T233859.SAFE', 'S2B_MSIL2A_20210627T100559_N0300_R022_T32TPS_20210627T135856.SAFE'], dtype='<U65') - eo:cloud_cover(time)float6412.65 14.92 16.5 ... 16.35 11.91
array([12.645555, 14.923827, 16.502707, 15.246907, 57.402623, 47.756503, 44.72053 , 56.068999, 96.218115, 92.314465, 62.109846, 55.815224, 21.750824, 24.577105, 5.444319, 5.673918, 35.295203, 27.991417, 97.720245, 96.397674, 26.698235, 20.468445, 36.142011, 44.22752 , 16.351688, 11.912692]) - instruments()<U3'msi'
array('msi', dtype='<U3') - s2:datatake_id(time)<U34'GS2B_20210528T100559_022070_N05...
array(['GS2B_20210528T100559_022070_N05.00', 'GS2B_20210528T100559_022070_N03.00', 'GS2B_20210531T101559_022113_N05.00', 'GS2B_20210531T101559_022113_N03.00', 'GS2A_20210602T101031_031050_N05.00', 'GS2A_20210602T101031_031050_N03.00', 'GS2A_20210605T102021_031093_N03.00', 'GS2A_20210605T102021_031093_N05.00', 'GS2B_20210607T100559_022213_N05.00', 'GS2B_20210607T100559_022213_N03.00', 'GS2B_20210610T101559_022256_N05.00', 'GS2B_20210610T101559_022256_N03.00', 'GS2A_20210612T101031_031193_N05.00', 'GS2A_20210612T101031_031193_N03.00', 'GS2A_20210615T102021_031236_N03.00', 'GS2A_20210615T102021_031236_N05.00', 'GS2B_20210617T100559_022356_N05.00', 'GS2B_20210617T100559_022356_N03.00', 'GS2B_20210620T101559_022399_N03.00', 'GS2B_20210620T101559_022399_N05.00', 'GS2A_20210622T101031_031336_N05.00', 'GS2A_20210622T101031_031336_N03.00', 'GS2A_20210625T102021_031379_N03.00', 'GS2A_20210625T102021_031379_N05.00', 'GS2B_20210627T100559_022499_N05.00', 'GS2B_20210627T100559_022499_N03.00'], dtype='<U34') - s2:nodata_pixel_percentage(time)object0 7e-06 11.874377 ... 0 7e-06
array([0, 7e-06, 11.874377, 11.646823, 0, 3e-06, 10.985302, 11.013988, 0, 0, 11.783398, 11.556501, 8.3e-05, 1.3e-05, 10.792472, 10.827903, 0, 1.7e-05, 11.396532, 11.627065, 0.000146, 1.7e-05, 10.879045, 10.92066, 0, 7e-06], dtype=object) - view:sun_azimuth(time)float64151.1 151.1 155.4 ... 146.8 146.8
array([151.07526534, 151.07388572, 155.39203616, 155.39059825, 150.16382152, 150.16244266, 154.53346131, 154.5349045 , 149.27864414, 149.2772675 , 153.70866815, 153.70724073, 148.4554036 , 148.45405463, 152.94501975, 152.94641223, 147.75003773, 147.74872101, 152.3076782 , 152.30905816, 147.19176534, 147.19046035, 151.8233914 , 151.82473308, 146.81010023, 146.80884882]) - earthsearch:s3_path(time)<U79's3://sentinel-cogs/sentinel-s2-...
array(['s3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/5/S2B_32TPS_20210528_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/5/S2B_32TPS_20210528_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/5/S2B_32TPS_20210531_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/5/S2B_32TPS_20210531_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210602_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210602_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210605_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210605_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210607_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210607_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210610_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210610_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210612_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210612_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210615_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210615_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210617_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210617_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210620_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210620_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210622_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210622_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210625_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210625_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210627_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210627_0_L2A'], dtype='<U79') - constellation()<U10'sentinel-2'
array('sentinel-2', dtype='<U10') - s2:granule_id(time)<U62'S2B_OPER_MSI_L2A_TL_S2RP_202302...
array(['S2B_OPER_MSI_L2A_TL_S2RP_20230227T082312_A022070_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20210528T130836_A022070_T32TPS_N03.00', 'S2B_OPER_MSI_L2A_TL_S2RP_20230301T002441_A022113_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS4_20210531T140040_A022113_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20230313T040411_A031050_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210602T171307_A031050_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210605T152329_A031093_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20230312T162726_A031093_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_S2RP_20230314T071412_A022213_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20210607T163734_A022213_T32TPS_N03.00', 'S2B_OPER_MSI_L2A_TL_S2RP_20230321T102457_A022256_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20210610T132739_A022256_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20230316T113702_A031193_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210612T131842_A031193_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210615T131659_A031236_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20230321T221050_A031236_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_S2RP_20230321T221728_A022356_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20210617T131219_A022356_T32TPS_N03.00', 'S2B_OPER_MSI_L2A_TL_VGS4_20210620T140552_A022399_T32TPS_N03.00', 'S2B_OPER_MSI_L2A_TL_S2RP_20230308T170500_A022399_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20230129T153031_A031336_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS4_20210622T121305_A031336_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210625T132502_A031379_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20230319T054911_A031379_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_S2RP_20230318T233859_A022499_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS4_20210627T135856_A022499_T32TPS_N03.00'], dtype='<U62') - s2:reflectance_conversion_factor(time)float640.975 0.975 0.974 ... 0.9679 0.9679
array([0.97504789, 0.97504789, 0.97399904, 0.97399904, 0.97334113, 0.97334113, 0.97240967, 0.97240967, 0.97183347, 0.97183347, 0.97102521, 0.97102521, 0.97053088, 0.97053088, 0.96985123, 0.96985123, 0.969445 , 0.969445 , 0.96889809, 0.96889809, 0.96858002, 0.96858002, 0.96816954, 0.96816954, 0.96794362, 0.96794362]) - grid:code()<U10'MGRS-32TPS'
array('MGRS-32TPS', dtype='<U10') - updated(time)<U24'2023-07-19T01:31:45.362Z' ... '...
array(['2023-07-19T01:31:45.362Z', '2022-11-06T13:34:17.157Z', '2023-08-21T02:48:42.746Z', '2022-11-06T04:43:15.876Z', '2023-07-14T10:06:11.004Z', '2022-11-06T04:43:21.072Z', '2022-11-06T04:42:40.826Z', '2023-08-19T18:25:04.412Z', '2023-07-15T20:10:05.181Z', '2022-11-06T04:44:31.882Z', '2023-08-28T22:22:56.940Z', '2022-11-06T04:43:35.373Z', '2023-08-31T14:25:17.439Z', '2022-11-06T04:44:47.117Z', '2022-11-06T04:44:42.815Z', '2023-08-28T20:41:39.160Z', '2023-08-28T19:50:44.246Z', '2022-11-06T04:43:20.506Z', '2022-11-06T04:46:02.240Z', '2023-07-16T11:32:44.388Z', '2023-07-12T22:29:59.130Z', '2022-11-06T04:46:04.928Z', '2022-11-06T04:45:30.338Z', '2023-09-01T04:30:40.825Z', '2023-09-02T00:59:09.859Z', '2022-11-06T04:45:27.038Z'], dtype='<U24') - created(time)<U24'2023-07-19T01:31:45.362Z' ... '...
array(['2023-07-19T01:31:45.362Z', '2022-11-03T10:53:30.936Z', '2023-08-21T02:48:42.746Z', '2022-11-06T04:43:15.876Z', '2023-07-14T10:06:11.004Z', '2022-11-06T04:43:21.072Z', '2022-11-06T04:42:40.826Z', '2023-08-19T18:25:04.412Z', '2023-07-15T20:10:05.181Z', '2022-11-06T04:44:31.882Z', '2023-08-28T22:22:56.940Z', '2022-11-06T04:43:35.373Z', '2023-08-31T14:25:17.439Z', '2022-11-06T04:44:47.117Z', '2022-11-06T04:44:42.815Z', '2023-08-28T20:41:39.160Z', '2023-08-28T19:50:44.246Z', '2022-11-06T04:43:20.506Z', '2022-11-06T04:46:02.240Z', '2023-07-16T11:32:44.388Z', '2023-07-12T22:29:59.130Z', '2022-11-06T04:46:04.928Z', '2022-11-06T04:45:30.338Z', '2023-09-01T04:30:40.825Z', '2023-09-02T00:59:09.859Z', '2022-11-06T04:45:27.038Z'], dtype='<U24') - view:sun_elevation(time)float6462.68 62.67 63.84 ... 63.8 63.79
array([62.67508338, 62.67485352, 63.83829126, 63.83809076, 63.2776855 , 63.27744956, 64.36252912, 64.36273616, 63.71164562, 63.71140388, 64.71725324, 64.71704181, 63.97586903, 63.9756252 , 64.90059226, 64.90080535, 64.07538254, 64.07513775, 64.91877022, 64.91898737, 64.01302019, 64.0127717 , 64.77526927, 64.77548806, 63.79508809, 63.79484238]) - s2:thin_cirrus_percentage(time)float6410.13 10.17 ... 0.6973 0.4268
array([10.13488 , 10.168401, 0.115349, 0.120899, 0.178427, 0.169714, 0.488913, 0.661111, 0.769374, 0.335158, 3.120367, 3.627113, 7.935338, 8.003794, 0.022728, 0.463668, 0.028782, 0.037817, 0.721744, 0.684867, 0.146924, 0.19993 , 0.375375, 0.267541, 0.697274, 0.426833]) - platform(time)<U11'sentinel-2b' ... 'sentinel-2b'
array(['sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b'], dtype='<U11') - s2:water_percentage(time)object0.20837 0.197246 ... 0.337965
array([0.20837, 0.197246, 0.271758, 0.382846, 0.142355, 0.298234, 0.443727, 0.187946, 0.022512, 0.027246, 0.175192, 0.226552, 0.209528, 0.239395, 0.357188, 0.292813, 0.272607, 0.497115, 0, 0.000169, 0.233775, 0.335679, 0.405969, 0.275791, 0.281174, 0.337965], dtype=object) - s2:dark_features_percentage(time)float640.9701 0.6094 ... 0.6925 0.9415
array([9.701360e-01, 6.094340e-01, 2.015643e+00, 1.392244e+00, 3.438870e-01, 6.917930e-01, 8.403930e-01, 1.378928e+00, 1.390000e-03, 4.481100e-02, 1.882207e+00, 7.810120e-01, 8.046860e-01, 5.937070e-01, 7.546780e-01, 1.151714e+00, 5.513320e-01, 7.116930e-01, 2.617000e-03, 2.910000e-03, 4.811240e-01, 9.703290e-01, 1.271975e+00, 9.746800e-01, 6.925060e-01, 9.414770e-01]) - s2:not_vegetated_percentage(time)float6414.25 9.691 7.788 ... 8.512 9.013
array([14.248499, 9.690771, 7.787908, 7.603369, 3.319488, 3.499338, 2.859635, 2.836192, 0.388685, 0.260192, 2.894815, 2.051123, 8.5719 , 6.641897, 9.223264, 9.58168 , 4.644649, 4.95012 , 0.113843, 1.005147, 8.066355, 9.131461, 4.460402, 3.650089, 8.511797, 9.012622]) - s2:saturated_defective_pixel_percentage()int640
array(0)
- s2:snow_ice_percentage(time)float6431.3 27.28 27.47 ... 5.971 6.136
array([31.303981, 27.278802, 27.466634, 24.24368 , 8.633302, 12.746431, 12.460077, 5.774973, 0.34478 , 3.110733, 6.592238, 9.930623, 19.589281, 15.464897, 16.500069, 19.468927, 9.188948, 10.062376, 0.083632, 1.050132, 6.899765, 7.237989, 3.306192, 1.222734, 5.971161, 6.136174]) - mgrs:grid_square()<U2'PS'
array('PS', dtype='<U2') - s2:cloud_shadow_percentage(time)object0.123971 0.265839 ... 1.637513
array([0.123971, 0.265839, 1.426617, 1.504799, 4.112166, 2.610499, 4.382209, 3.633687, 0.766328, 0.140311, 1.309796, 2.989328, 0.735473, 0.90655, 0.556963, 0.533688, 3.479381, 3.173427, 0, 0.013745, 5.264171, 2.58029, 4.672229, 5.893223, 2.496737, 1.637513], dtype=object) - mgrs:utm_zone()int6432
array(32)
- s2:high_proba_clouds_percentage(time)float641.311 1.87 11.34 ... 8.549 9.308
array([ 1.310789, 1.869626, 11.341699, 11.836569, 41.895783, 42.292377, 37.297574, 37.018588, 78.666943, 79.059446, 45.465553, 45.84513 , 5.113235, 5.835326, 4.018103, 2.9164 , 23.718023, 24.355602, 27.910239, 27.134106, 16.794626, 17.557459, 31.651893, 31.285802, 8.549391, 9.307959]) - earthsearch:boa_offset_applied(time)boolTrue False True ... True True False
array([ True, False, True, False, True, False, False, True, True, False, True, False, True, False, False, True, True, False, False, True, True, False, False, True, True, False]) - s2:medium_proba_clouds_percentage(time)float641.2 2.886 5.046 ... 7.105 2.178
array([ 1.199887, 2.8858 , 5.045658, 3.289439, 15.328416, 5.294412, 6.934044, 18.3893 , 16.781795, 12.919861, 13.523927, 6.342981, 8.702251, 10.737985, 1.403487, 2.29385 , 11.548399, 3.597998, 69.088262, 68.578702, 9.756684, 2.711056, 4.114743, 12.67418 , 7.105023, 2.177899]) - s2:vegetation_percentage(time)float6440.25 40.69 44.01 ... 65.32 67.63
array([40.248781, 40.689981, 44.01297 , 45.765302, 24.552888, 27.177703, 30.178928, 28.761679, 1.449043, 2.117007, 22.656791, 23.482373, 46.2971 , 47.072074, 64.543813, 63.144523, 45.910549, 48.674297, 1.309192, 1.238064, 51.928717, 56.201601, 46.205747, 42.820308, 65.318006, 67.6287 ]) - s2:degraded_msi_data_percentage(time)object0.0112 0 0.0265 ... 0.1149 0.0127 0
array([0.0112, 0, 0.0265, 0, 0.1008, 0, 0, 0.2234, 0.1724, 0, 0.0443, 0, 0.0044, 0, 0, 0.0258, 0.0333, 0, 0, 0.0306, 0.0309, 0, 0, 0.1149, 0.0127, 0], dtype=object) - earthsearch:payload_id(time)<U74'roda-sentinel2/workflow-sentine...
array(['roda-sentinel2/workflow-sentinel2-to-stac/b84ebd104933e25116f9d2b158090bad', 'roda-sentinel2/workflow-sentinel2-to-stac/4565558a179b902dbd75791ba0821c05', 'roda-sentinel2/workflow-sentinel2-to-stac/9a9188736d6ffe2307874ac719c17178', 'roda-sentinel2/workflow-sentinel2-to-stac/fa9c01876d73bd7d39c8229cf48b4798', 'roda-sentinel2/workflow-sentinel2-to-stac/24be754211f90b660f6f3f82405e8c20', 'roda-sentinel2/workflow-sentinel2-to-stac/aa59d15d1c3bb88a838de30d9bca323a', 'roda-sentinel2/workflow-sentinel2-to-stac/c6b8893f8709d604a4501d85cae0ff1d', 'roda-sentinel2/workflow-sentinel2-to-stac/c2d5ccf5f168e5f3be4b6a420366f272', 'roda-sentinel2/workflow-sentinel2-to-stac/e7007d1cc5a002936cc6b3f3455f25d2', 'roda-sentinel2/workflow-sentinel2-to-stac/d8d561f2e1fddceff644293edac5a973', 'roda-sentinel2/workflow-sentinel2-to-stac/bfebbdc9c825add7648ced2d0f4795af', 'roda-sentinel2/workflow-sentinel2-to-stac/f806c3f35353dd870127f2df07aa0791', 'roda-sentinel2/workflow-sentinel2-to-stac/b156ec150b4550cf8b9d8456b9078007', 'roda-sentinel2/workflow-sentinel2-to-stac/2d850f82bac7b2a3b7d5daaade4ec1fc', 'roda-sentinel2/workflow-sentinel2-to-stac/8f4ef169c9800d925e7b78ed83b6243c', 'roda-sentinel2/workflow-sentinel2-to-stac/9a5972ef4da914562d2d2513790d027b', 'roda-sentinel2/workflow-sentinel2-to-stac/0318dd97ab3148f08142ef2bd1793c90', 'roda-sentinel2/workflow-sentinel2-to-stac/17239972afbdaaebb2408a7e0ebd5823', 'roda-sentinel2/workflow-sentinel2-to-stac/e0d4e2a19db22497be3d3ab319e5dd0a', 'roda-sentinel2/workflow-sentinel2-to-stac/d55883951e691ff8016328ad13ba5a80', 'roda-sentinel2/workflow-sentinel2-to-stac/ed7567a4bf3a0704cf8c35c32a534284', 'roda-sentinel2/workflow-sentinel2-to-stac/fde1a94a72e701b06ca16e3203c98fcc', 'roda-sentinel2/workflow-sentinel2-to-stac/98c0711e502f7af7cef9d582c2c5d4b1', 'roda-sentinel2/workflow-sentinel2-to-stac/9dc06d10fd3eef6b7f01c000dce4a184', 'roda-sentinel2/workflow-sentinel2-to-stac/39a848b9c950e88f80c1f5529b45d49a', 'roda-sentinel2/workflow-sentinel2-to-stac/c796a8ece534e903bb7d58398b39f859'], dtype='<U74') - gsd()int6410
array(10)
- proj:shape()object{10980}
array({10980}, dtype=object) - proj:transform()object{0, 600000, 10, 5200020, -10}
array({0, 600000, 10, 5200020, -10}, dtype=object) - title(band)<U20'Red (band 4) - 10m' 'NIR 1 (ban...
array(['Red (band 4) - 10m', 'NIR 1 (band 8) - 10m'], dtype='<U20')
- common_name(band)<U3'red' 'nir'
array(['red', 'nir'], dtype='<U3')
- center_wavelength(band)float640.665 0.842
array([0.665, 0.842])
- full_width_half_max(band)float640.038 0.145
array([0.038, 0.145])
- epsg()int6432632
array(32632)
- timePandasIndex
PandasIndex(DatetimeIndex(['2021-05-28 10:17:59.882000', '2021-05-28 10:17:59.884000', '2021-05-31 10:27:56.286000', '2021-05-31 10:27:56.289000', '2021-06-02 10:18:00.103000', '2021-06-02 10:18:00.104000', '2021-06-05 10:27:56.280000', '2021-06-05 10:27:56.282000', '2021-06-07 10:18:00.323000', '2021-06-07 10:18:00.323000', '2021-06-10 10:27:56.565000', '2021-06-10 10:27:56.566000', '2021-06-12 10:17:59.823000', '2021-06-12 10:17:59.824000', '2021-06-15 10:27:55.829000', '2021-06-15 10:27:55.830000', '2021-06-17 10:18:00.191000', '2021-06-17 10:18:00.192000', '2021-06-20 10:27:56.235000', '2021-06-20 10:27:56.236000', '2021-06-22 10:18:00.204000', '2021-06-22 10:18:00.205000', '2021-06-25 10:27:56.865000', '2021-06-25 10:27:56.867000', '2021-06-27 10:18:00.077000', '2021-06-27 10:18:00.078000'], dtype='datetime64[ns]', name='time', freq=None)) - bandPandasIndex
PandasIndex(Index(['red', 'nir'], dtype='object', name='band'))
- xPandasIndex
PandasIndex(Index([673310.0, 673320.0, 673330.0, 673340.0, 673350.0, 673360.0, 673370.0, 673380.0, 673390.0, 673400.0, ... 684670.0, 684680.0, 684690.0, 684700.0, 684710.0, 684720.0, 684730.0, 684740.0, 684750.0, 684760.0], dtype='float64', name='x', length=1146)) - yPandasIndex
PandasIndex(Index([5154890.0, 5154880.0, 5154870.0, 5154860.0, 5154850.0, 5154840.0, 5154830.0, 5154820.0, 5154810.0, 5154800.0, ... 5147850.0, 5147840.0, 5147830.0, 5147820.0, 5147810.0, 5147800.0, 5147790.0, 5147780.0, 5147770.0, 5147760.0], dtype='float64', name='y', length=714))
- spec :
- RasterSpec(epsg=32632, bounds=(673310.0, 5147750.0, 684770.0, 5154890.0), resolutions_xy=(10.0, 10.0))
- crs :
- epsg:32632
- transform :
- | 10.00, 0.00, 673310.00| | 0.00,-10.00, 5154890.00| | 0.00, 0.00, 1.00|
- resolution :
- 10.0
Get rid of possible negative values#
s2_cube = xr.where(s2_cube>=0, s2_cube, 0)
Reduce the time dimension by averaging along the time dimension. Check what happens to the datacube inspecting the resulting xArray object:
datacube_mean_time = s2_cube.mean("time")
datacube_mean_time
<xarray.DataArray 'stackstac-b671372a6175b16d2e4eaf48f5606039' (band: 2,
y: 714, x: 1146)>
dask.array<mean_agg-aggregate, shape=(2, 714, 1146), dtype=float64, chunksize=(1, 714, 1024), chunktype=numpy.ndarray>
Coordinates: (12/21)
* band (band) <U3 'red' 'nir'
* x (x) float64 6.733e+05 ... 6.848e+05
* y (y) float64 5.155e+06 ... 5.148e+06
s2:product_type <U7 'S2MSI2A'
s2:datatake_type <U8 'INS-NOBS'
proj:epsg int64 32632
... ...
proj:transform object {0, 600000, 10, 5200020, ...
title (band) <U20 'Red (band 4) - 10m'...
common_name (band) <U3 'red' 'nir'
center_wavelength (band) float64 0.665 0.842
full_width_half_max (band) float64 0.038 0.145
epsg int64 32632- band: 2
- y: 714
- x: 1146
- dask.array<chunksize=(1, 714, 1024), meta=np.ndarray>
Array Chunk Bytes 12.49 MiB 5.58 MiB Shape (2, 714, 1146) (1, 714, 1024) Dask graph 4 chunks in 9 graph layers Data type float64 numpy.ndarray - band(band)<U3'red' 'nir'
array(['red', 'nir'], dtype='<U3')
- x(x)float646.733e+05 6.733e+05 ... 6.848e+05
array([673310., 673320., 673330., ..., 684740., 684750., 684760.])
- y(y)float645.155e+06 5.155e+06 ... 5.148e+06
array([5154890., 5154880., 5154870., ..., 5147780., 5147770., 5147760.])
- s2:product_type()<U7'S2MSI2A'
array('S2MSI2A', dtype='<U7') - s2:datatake_type()<U8'INS-NOBS'
array('INS-NOBS', dtype='<U8') - proj:epsg()int6432632
array(32632)
- mgrs:latitude_band()<U1'T'
array('T', dtype='<U1') - instruments()<U3'msi'
array('msi', dtype='<U3') - constellation()<U10'sentinel-2'
array('sentinel-2', dtype='<U10') - grid:code()<U10'MGRS-32TPS'
array('MGRS-32TPS', dtype='<U10') - s2:saturated_defective_pixel_percentage()int640
array(0)
- mgrs:grid_square()<U2'PS'
array('PS', dtype='<U2') - mgrs:utm_zone()int6432
array(32)
- gsd()int6410
array(10)
- proj:shape()object{10980}
array({10980}, dtype=object) - proj:transform()object{0, 600000, 10, 5200020, -10}
array({0, 600000, 10, 5200020, -10}, dtype=object) - title(band)<U20'Red (band 4) - 10m' 'NIR 1 (ban...
array(['Red (band 4) - 10m', 'NIR 1 (band 8) - 10m'], dtype='<U20')
- common_name(band)<U3'red' 'nir'
array(['red', 'nir'], dtype='<U3')
- center_wavelength(band)float640.665 0.842
array([0.665, 0.842])
- full_width_half_max(band)float640.038 0.145
array([0.038, 0.145])
- epsg()int6432632
array(32632)
- bandPandasIndex
PandasIndex(Index(['red', 'nir'], dtype='object', name='band'))
- xPandasIndex
PandasIndex(Index([673310.0, 673320.0, 673330.0, 673340.0, 673350.0, 673360.0, 673370.0, 673380.0, 673390.0, 673400.0, ... 684670.0, 684680.0, 684690.0, 684700.0, 684710.0, 684720.0, 684730.0, 684740.0, 684750.0, 684760.0], dtype='float64', name='x', length=1146)) - yPandasIndex
PandasIndex(Index([5154890.0, 5154880.0, 5154870.0, 5154860.0, 5154850.0, 5154840.0, 5154830.0, 5154820.0, 5154810.0, 5154800.0, ... 5147850.0, 5147840.0, 5147830.0, 5147820.0, 5147810.0, 5147800.0, 5147790.0, 5147780.0, 5147770.0, 5147760.0], dtype='float64', name='y', length=714))
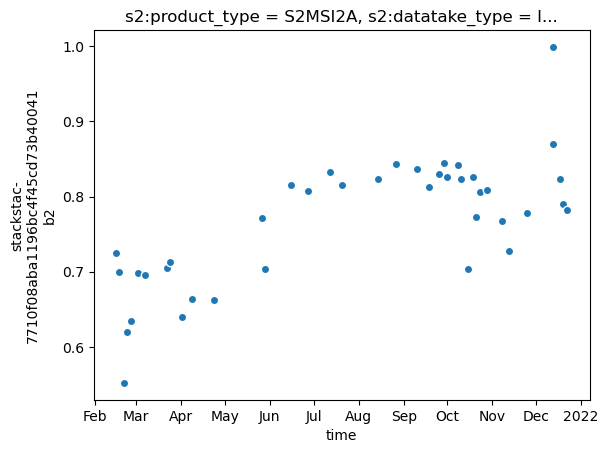
It is possible to reduce in the same way all the available dimensions of the datacube.
We can, for instance, reduce the band dimension similarly as we did for the temporal dimension:
The result will now contain values resulting from the average of the bands:
datacube_mean_band = s2_cube.mean("band")
datacube_mean_band
<xarray.DataArray 'stackstac-b671372a6175b16d2e4eaf48f5606039' (time: 26,
y: 714, x: 1146)>
dask.array<mean_agg-aggregate, shape=(26, 714, 1146), dtype=float64, chunksize=(1, 714, 1024), chunktype=numpy.ndarray>
Coordinates: (12/48)
* time (time) datetime64[ns] 2021-05-28...
id (time) <U24 'S2B_32TPS_20210528_...
* x (x) float64 6.733e+05 ... 6.848e+05
* y (y) float64 5.155e+06 ... 5.148e+06
s2:unclassified_percentage (time) float64 0.2507 ... 2.393
s2:product_type <U7 'S2MSI2A'
... ...
s2:degraded_msi_data_percentage (time) object 0.0112 0 ... 0.0127 0
earthsearch:payload_id (time) <U74 'roda-sentinel2/work...
gsd int64 10
proj:shape object {10980}
proj:transform object {0, 600000, 10, 5200020, ...
epsg int64 32632- time: 26
- y: 714
- x: 1146
- dask.array<chunksize=(1, 714, 1024), meta=np.ndarray>
Array Chunk Bytes 162.31 MiB 5.58 MiB Shape (26, 714, 1146) (1, 714, 1024) Dask graph 52 chunks in 7 graph layers Data type float64 numpy.ndarray - time(time)datetime64[ns]2021-05-28T10:17:59.882000 ... 2...
array(['2021-05-28T10:17:59.882000000', '2021-05-28T10:17:59.884000000', '2021-05-31T10:27:56.286000000', '2021-05-31T10:27:56.289000000', '2021-06-02T10:18:00.103000000', '2021-06-02T10:18:00.104000000', '2021-06-05T10:27:56.280000000', '2021-06-05T10:27:56.282000000', '2021-06-07T10:18:00.323000000', '2021-06-07T10:18:00.323000000', '2021-06-10T10:27:56.565000000', '2021-06-10T10:27:56.566000000', '2021-06-12T10:17:59.823000000', '2021-06-12T10:17:59.824000000', '2021-06-15T10:27:55.829000000', '2021-06-15T10:27:55.830000000', '2021-06-17T10:18:00.191000000', '2021-06-17T10:18:00.192000000', '2021-06-20T10:27:56.235000000', '2021-06-20T10:27:56.236000000', '2021-06-22T10:18:00.204000000', '2021-06-22T10:18:00.205000000', '2021-06-25T10:27:56.865000000', '2021-06-25T10:27:56.867000000', '2021-06-27T10:18:00.077000000', '2021-06-27T10:18:00.078000000'], dtype='datetime64[ns]') - id(time)<U24'S2B_32TPS_20210528_1_L2A' ... '...
array(['S2B_32TPS_20210528_1_L2A', 'S2B_32TPS_20210528_0_L2A', 'S2B_32TPS_20210531_1_L2A', 'S2B_32TPS_20210531_0_L2A', 'S2A_32TPS_20210602_1_L2A', 'S2A_32TPS_20210602_0_L2A', 'S2A_32TPS_20210605_0_L2A', 'S2A_32TPS_20210605_1_L2A', 'S2B_32TPS_20210607_1_L2A', 'S2B_32TPS_20210607_0_L2A', 'S2B_32TPS_20210610_1_L2A', 'S2B_32TPS_20210610_0_L2A', 'S2A_32TPS_20210612_1_L2A', 'S2A_32TPS_20210612_0_L2A', 'S2A_32TPS_20210615_0_L2A', 'S2A_32TPS_20210615_1_L2A', 'S2B_32TPS_20210617_1_L2A', 'S2B_32TPS_20210617_0_L2A', 'S2B_32TPS_20210620_0_L2A', 'S2B_32TPS_20210620_1_L2A', 'S2A_32TPS_20210622_1_L2A', 'S2A_32TPS_20210622_0_L2A', 'S2A_32TPS_20210625_0_L2A', 'S2A_32TPS_20210625_1_L2A', 'S2B_32TPS_20210627_1_L2A', 'S2B_32TPS_20210627_0_L2A'], dtype='<U24') - x(x)float646.733e+05 6.733e+05 ... 6.848e+05
array([673310., 673320., 673330., ..., 684740., 684750., 684760.])
- y(y)float645.155e+06 5.155e+06 ... 5.148e+06
array([5154890., 5154880., 5154870., ..., 5147780., 5147770., 5147760.])
- s2:unclassified_percentage(time)float640.2507 6.344 ... 0.3769 2.393
array([0.250706, 6.3441 , 0.515761, 3.860853, 1.493286, 5.219495, 4.114496, 1.357601, 0.809151, 1.985232, 2.379116, 4.723761, 2.041206, 4.504375, 2.619704, 0.152735, 0.65733 , 3.93955 , 0.770469, 0.292153, 0.427856, 3.074201, 3.535479, 0.935654, 0.376936, 2.392862]) - s2:product_type()<U7'S2MSI2A'
array('S2MSI2A', dtype='<U7') - s2:sequence(time)<U1'1' '0' '1' '0' ... '0' '1' '1' '0'
array(['1', '0', '1', '0', '1', '0', '0', '1', '1', '0', '1', '0', '1', '0', '0', '1', '1', '0', '0', '1', '1', '0', '0', '1', '1', '0'], dtype='<U1') - s2:generation_time(time)<U27'2023-02-27T08:23:12.000000Z' .....
array(['2023-02-27T08:23:12.000000Z', '2021-05-28T13:08:36.000000Z', '2023-03-01T00:24:41.000000Z', '2021-05-31T14:00:40.000000Z', '2023-03-13T04:04:11.000000Z', '2021-06-02T17:13:07.000000Z', '2021-06-05T15:23:29.000000Z', '2023-03-12T16:27:26.000000Z', '2023-03-14T07:14:12.000000Z', '2021-06-07T16:37:34.000000Z', '2023-03-21T10:24:57.000000Z', '2021-06-10T13:27:39.000000Z', '2023-03-16T11:37:02.000000Z', '2021-06-12T13:18:42.000000Z', '2021-06-15T13:16:59.000000Z', '2023-03-21T22:10:50.000000Z', '2023-03-21T22:17:28.000000Z', '2021-06-17T13:12:19.000000Z', '2021-06-20T14:05:52.000000Z', '2023-03-08T17:05:00.000000Z', '2023-01-29T15:30:31.000000Z', '2021-06-22T12:13:05.000000Z', '2021-06-25T13:25:02.000000Z', '2023-03-19T05:49:11.000000Z', '2023-03-18T23:38:59.000000Z', '2021-06-27T13:58:56.000000Z'], dtype='<U27') - s2:datatake_type()<U8'INS-NOBS'
array('INS-NOBS', dtype='<U8') - proj:epsg()int6432632
array(32632)
- mgrs:latitude_band()<U1'T'
array('T', dtype='<U1') - processing:software(time)object{'sentinel2-to-stac': '0.1.1'} ....
array([{'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}], dtype=object) - s2:processing_baseline(time)<U5'05.00' '03.00' ... '05.00' '03.00'
array(['05.00', '03.00', '05.00', '03.00', '05.00', '03.00', '03.00', '05.00', '05.00', '03.00', '05.00', '03.00', '05.00', '03.00', '03.00', '05.00', '05.00', '03.00', '03.00', '05.00', '05.00', '03.00', '03.00', '05.00', '05.00', '03.00'], dtype='<U5') - s2:datastrip_id(time)<U64'S2B_OPER_MSI_L2A_DS_S2RP_202302...
array(['S2B_OPER_MSI_L2A_DS_S2RP_20230227T082312_S20210528T100555_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20210528T130836_S20210528T100555_N03.00', 'S2B_OPER_MSI_L2A_DS_S2RP_20230301T002441_S20210531T102618_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS4_20210531T140040_S20210531T102618_N03.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20230313T040411_S20210602T101235_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210602T171307_S20210602T101235_N03.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210605T152329_S20210605T102737_N03.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20230312T162726_S20210605T102737_N05.00', 'S2B_OPER_MSI_L2A_DS_S2RP_20230314T071412_S20210607T101225_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20210607T163734_S20210607T101225_N03.00', 'S2B_OPER_MSI_L2A_DS_S2RP_20230321T102457_S20210610T102220_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20210610T132739_S20210610T102220_N03.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20230316T113702_S20210612T101329_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210612T131842_S20210612T101329_N03.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210615T131659_S20210615T102602_N03.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20230321T221050_S20210615T102602_N05.00', 'S2B_OPER_MSI_L2A_DS_S2RP_20230321T221728_S20210617T101301_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20210617T131219_S20210617T101301_N03.00', 'S2B_OPER_MSI_L2A_DS_VGS4_20210620T140552_S20210620T102722_N03.00', 'S2B_OPER_MSI_L2A_DS_S2RP_20230308T170500_S20210620T102722_N05.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20230129T153031_S20210622T101416_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS4_20210622T121305_S20210622T101416_N03.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210625T132502_S20210625T102545_N03.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20230319T054911_S20210625T102545_N05.00', 'S2B_OPER_MSI_L2A_DS_S2RP_20230318T233859_S20210627T101702_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS4_20210627T135856_S20210627T101702_N03.00'], dtype='<U64') - s2:product_uri(time)<U65'S2B_MSIL2A_20210528T100559_N050...
array(['S2B_MSIL2A_20210528T100559_N0500_R022_T32TPS_20230227T082312.SAFE', 'S2B_MSIL2A_20210528T100559_N0300_R022_T32TPS_20210528T130836.SAFE', 'S2B_MSIL2A_20210531T101559_N0500_R065_T32TPS_20230301T002441.SAFE', 'S2B_MSIL2A_20210531T101559_N0300_R065_T32TPS_20210531T140040.SAFE', 'S2A_MSIL2A_20210602T101031_N0500_R022_T32TPS_20230313T040411.SAFE', 'S2A_MSIL2A_20210602T101031_N0300_R022_T32TPS_20210602T171307.SAFE', 'S2A_MSIL2A_20210605T102021_N0300_R065_T32TPS_20210605T152329.SAFE', 'S2A_MSIL2A_20210605T102021_N0500_R065_T32TPS_20230312T162726.SAFE', 'S2B_MSIL2A_20210607T100559_N0500_R022_T32TPS_20230314T071412.SAFE', 'S2B_MSIL2A_20210607T100559_N0300_R022_T32TPS_20210607T163734.SAFE', 'S2B_MSIL2A_20210610T101559_N0500_R065_T32TPS_20230321T102457.SAFE', 'S2B_MSIL2A_20210610T101559_N0300_R065_T32TPS_20210610T132739.SAFE', 'S2A_MSIL2A_20210612T101031_N0500_R022_T32TPS_20230316T113702.SAFE', 'S2A_MSIL2A_20210612T101031_N0300_R022_T32TPS_20210612T131842.SAFE', 'S2A_MSIL2A_20210615T102021_N0300_R065_T32TPS_20210615T131659.SAFE', 'S2A_MSIL2A_20210615T102021_N0500_R065_T32TPS_20230321T221050.SAFE', 'S2B_MSIL2A_20210617T100559_N0500_R022_T32TPS_20230321T221728.SAFE', 'S2B_MSIL2A_20210617T100559_N0300_R022_T32TPS_20210617T131219.SAFE', 'S2B_MSIL2A_20210620T101559_N0300_R065_T32TPS_20210620T140552.SAFE', 'S2B_MSIL2A_20210620T101559_N0500_R065_T32TPS_20230308T170500.SAFE', 'S2A_MSIL2A_20210622T101031_N0500_R022_T32TPS_20230129T153031.SAFE', 'S2A_MSIL2A_20210622T101031_N0300_R022_T32TPS_20210622T121305.SAFE', 'S2A_MSIL2A_20210625T102021_N0300_R065_T32TPS_20210625T132502.SAFE', 'S2A_MSIL2A_20210625T102021_N0500_R065_T32TPS_20230319T054911.SAFE', 'S2B_MSIL2A_20210627T100559_N0500_R022_T32TPS_20230318T233859.SAFE', 'S2B_MSIL2A_20210627T100559_N0300_R022_T32TPS_20210627T135856.SAFE'], dtype='<U65') - eo:cloud_cover(time)float6412.65 14.92 16.5 ... 16.35 11.91
array([12.645555, 14.923827, 16.502707, 15.246907, 57.402623, 47.756503, 44.72053 , 56.068999, 96.218115, 92.314465, 62.109846, 55.815224, 21.750824, 24.577105, 5.444319, 5.673918, 35.295203, 27.991417, 97.720245, 96.397674, 26.698235, 20.468445, 36.142011, 44.22752 , 16.351688, 11.912692]) - instruments()<U3'msi'
array('msi', dtype='<U3') - s2:datatake_id(time)<U34'GS2B_20210528T100559_022070_N05...
array(['GS2B_20210528T100559_022070_N05.00', 'GS2B_20210528T100559_022070_N03.00', 'GS2B_20210531T101559_022113_N05.00', 'GS2B_20210531T101559_022113_N03.00', 'GS2A_20210602T101031_031050_N05.00', 'GS2A_20210602T101031_031050_N03.00', 'GS2A_20210605T102021_031093_N03.00', 'GS2A_20210605T102021_031093_N05.00', 'GS2B_20210607T100559_022213_N05.00', 'GS2B_20210607T100559_022213_N03.00', 'GS2B_20210610T101559_022256_N05.00', 'GS2B_20210610T101559_022256_N03.00', 'GS2A_20210612T101031_031193_N05.00', 'GS2A_20210612T101031_031193_N03.00', 'GS2A_20210615T102021_031236_N03.00', 'GS2A_20210615T102021_031236_N05.00', 'GS2B_20210617T100559_022356_N05.00', 'GS2B_20210617T100559_022356_N03.00', 'GS2B_20210620T101559_022399_N03.00', 'GS2B_20210620T101559_022399_N05.00', 'GS2A_20210622T101031_031336_N05.00', 'GS2A_20210622T101031_031336_N03.00', 'GS2A_20210625T102021_031379_N03.00', 'GS2A_20210625T102021_031379_N05.00', 'GS2B_20210627T100559_022499_N05.00', 'GS2B_20210627T100559_022499_N03.00'], dtype='<U34') - s2:nodata_pixel_percentage(time)object0 7e-06 11.874377 ... 0 7e-06
array([0, 7e-06, 11.874377, 11.646823, 0, 3e-06, 10.985302, 11.013988, 0, 0, 11.783398, 11.556501, 8.3e-05, 1.3e-05, 10.792472, 10.827903, 0, 1.7e-05, 11.396532, 11.627065, 0.000146, 1.7e-05, 10.879045, 10.92066, 0, 7e-06], dtype=object) - view:sun_azimuth(time)float64151.1 151.1 155.4 ... 146.8 146.8
array([151.07526534, 151.07388572, 155.39203616, 155.39059825, 150.16382152, 150.16244266, 154.53346131, 154.5349045 , 149.27864414, 149.2772675 , 153.70866815, 153.70724073, 148.4554036 , 148.45405463, 152.94501975, 152.94641223, 147.75003773, 147.74872101, 152.3076782 , 152.30905816, 147.19176534, 147.19046035, 151.8233914 , 151.82473308, 146.81010023, 146.80884882]) - earthsearch:s3_path(time)<U79's3://sentinel-cogs/sentinel-s2-...
array(['s3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/5/S2B_32TPS_20210528_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/5/S2B_32TPS_20210528_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/5/S2B_32TPS_20210531_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/5/S2B_32TPS_20210531_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210602_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210602_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210605_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210605_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210607_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210607_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210610_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210610_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210612_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210612_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210615_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210615_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210617_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210617_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210620_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210620_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210622_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210622_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210625_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210625_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210627_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210627_0_L2A'], dtype='<U79') - constellation()<U10'sentinel-2'
array('sentinel-2', dtype='<U10') - s2:granule_id(time)<U62'S2B_OPER_MSI_L2A_TL_S2RP_202302...
array(['S2B_OPER_MSI_L2A_TL_S2RP_20230227T082312_A022070_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20210528T130836_A022070_T32TPS_N03.00', 'S2B_OPER_MSI_L2A_TL_S2RP_20230301T002441_A022113_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS4_20210531T140040_A022113_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20230313T040411_A031050_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210602T171307_A031050_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210605T152329_A031093_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20230312T162726_A031093_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_S2RP_20230314T071412_A022213_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20210607T163734_A022213_T32TPS_N03.00', 'S2B_OPER_MSI_L2A_TL_S2RP_20230321T102457_A022256_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20210610T132739_A022256_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20230316T113702_A031193_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210612T131842_A031193_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210615T131659_A031236_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20230321T221050_A031236_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_S2RP_20230321T221728_A022356_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20210617T131219_A022356_T32TPS_N03.00', 'S2B_OPER_MSI_L2A_TL_VGS4_20210620T140552_A022399_T32TPS_N03.00', 'S2B_OPER_MSI_L2A_TL_S2RP_20230308T170500_A022399_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20230129T153031_A031336_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS4_20210622T121305_A031336_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210625T132502_A031379_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20230319T054911_A031379_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_S2RP_20230318T233859_A022499_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS4_20210627T135856_A022499_T32TPS_N03.00'], dtype='<U62') - s2:reflectance_conversion_factor(time)float640.975 0.975 0.974 ... 0.9679 0.9679
array([0.97504789, 0.97504789, 0.97399904, 0.97399904, 0.97334113, 0.97334113, 0.97240967, 0.97240967, 0.97183347, 0.97183347, 0.97102521, 0.97102521, 0.97053088, 0.97053088, 0.96985123, 0.96985123, 0.969445 , 0.969445 , 0.96889809, 0.96889809, 0.96858002, 0.96858002, 0.96816954, 0.96816954, 0.96794362, 0.96794362]) - grid:code()<U10'MGRS-32TPS'
array('MGRS-32TPS', dtype='<U10') - updated(time)<U24'2023-07-19T01:31:45.362Z' ... '...
array(['2023-07-19T01:31:45.362Z', '2022-11-06T13:34:17.157Z', '2023-08-21T02:48:42.746Z', '2022-11-06T04:43:15.876Z', '2023-07-14T10:06:11.004Z', '2022-11-06T04:43:21.072Z', '2022-11-06T04:42:40.826Z', '2023-08-19T18:25:04.412Z', '2023-07-15T20:10:05.181Z', '2022-11-06T04:44:31.882Z', '2023-08-28T22:22:56.940Z', '2022-11-06T04:43:35.373Z', '2023-08-31T14:25:17.439Z', '2022-11-06T04:44:47.117Z', '2022-11-06T04:44:42.815Z', '2023-08-28T20:41:39.160Z', '2023-08-28T19:50:44.246Z', '2022-11-06T04:43:20.506Z', '2022-11-06T04:46:02.240Z', '2023-07-16T11:32:44.388Z', '2023-07-12T22:29:59.130Z', '2022-11-06T04:46:04.928Z', '2022-11-06T04:45:30.338Z', '2023-09-01T04:30:40.825Z', '2023-09-02T00:59:09.859Z', '2022-11-06T04:45:27.038Z'], dtype='<U24') - created(time)<U24'2023-07-19T01:31:45.362Z' ... '...
array(['2023-07-19T01:31:45.362Z', '2022-11-03T10:53:30.936Z', '2023-08-21T02:48:42.746Z', '2022-11-06T04:43:15.876Z', '2023-07-14T10:06:11.004Z', '2022-11-06T04:43:21.072Z', '2022-11-06T04:42:40.826Z', '2023-08-19T18:25:04.412Z', '2023-07-15T20:10:05.181Z', '2022-11-06T04:44:31.882Z', '2023-08-28T22:22:56.940Z', '2022-11-06T04:43:35.373Z', '2023-08-31T14:25:17.439Z', '2022-11-06T04:44:47.117Z', '2022-11-06T04:44:42.815Z', '2023-08-28T20:41:39.160Z', '2023-08-28T19:50:44.246Z', '2022-11-06T04:43:20.506Z', '2022-11-06T04:46:02.240Z', '2023-07-16T11:32:44.388Z', '2023-07-12T22:29:59.130Z', '2022-11-06T04:46:04.928Z', '2022-11-06T04:45:30.338Z', '2023-09-01T04:30:40.825Z', '2023-09-02T00:59:09.859Z', '2022-11-06T04:45:27.038Z'], dtype='<U24') - view:sun_elevation(time)float6462.68 62.67 63.84 ... 63.8 63.79
array([62.67508338, 62.67485352, 63.83829126, 63.83809076, 63.2776855 , 63.27744956, 64.36252912, 64.36273616, 63.71164562, 63.71140388, 64.71725324, 64.71704181, 63.97586903, 63.9756252 , 64.90059226, 64.90080535, 64.07538254, 64.07513775, 64.91877022, 64.91898737, 64.01302019, 64.0127717 , 64.77526927, 64.77548806, 63.79508809, 63.79484238]) - s2:thin_cirrus_percentage(time)float6410.13 10.17 ... 0.6973 0.4268
array([10.13488 , 10.168401, 0.115349, 0.120899, 0.178427, 0.169714, 0.488913, 0.661111, 0.769374, 0.335158, 3.120367, 3.627113, 7.935338, 8.003794, 0.022728, 0.463668, 0.028782, 0.037817, 0.721744, 0.684867, 0.146924, 0.19993 , 0.375375, 0.267541, 0.697274, 0.426833]) - platform(time)<U11'sentinel-2b' ... 'sentinel-2b'
array(['sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b'], dtype='<U11') - s2:water_percentage(time)object0.20837 0.197246 ... 0.337965
array([0.20837, 0.197246, 0.271758, 0.382846, 0.142355, 0.298234, 0.443727, 0.187946, 0.022512, 0.027246, 0.175192, 0.226552, 0.209528, 0.239395, 0.357188, 0.292813, 0.272607, 0.497115, 0, 0.000169, 0.233775, 0.335679, 0.405969, 0.275791, 0.281174, 0.337965], dtype=object) - s2:dark_features_percentage(time)float640.9701 0.6094 ... 0.6925 0.9415
array([9.701360e-01, 6.094340e-01, 2.015643e+00, 1.392244e+00, 3.438870e-01, 6.917930e-01, 8.403930e-01, 1.378928e+00, 1.390000e-03, 4.481100e-02, 1.882207e+00, 7.810120e-01, 8.046860e-01, 5.937070e-01, 7.546780e-01, 1.151714e+00, 5.513320e-01, 7.116930e-01, 2.617000e-03, 2.910000e-03, 4.811240e-01, 9.703290e-01, 1.271975e+00, 9.746800e-01, 6.925060e-01, 9.414770e-01]) - s2:not_vegetated_percentage(time)float6414.25 9.691 7.788 ... 8.512 9.013
array([14.248499, 9.690771, 7.787908, 7.603369, 3.319488, 3.499338, 2.859635, 2.836192, 0.388685, 0.260192, 2.894815, 2.051123, 8.5719 , 6.641897, 9.223264, 9.58168 , 4.644649, 4.95012 , 0.113843, 1.005147, 8.066355, 9.131461, 4.460402, 3.650089, 8.511797, 9.012622]) - s2:saturated_defective_pixel_percentage()int640
array(0)
- s2:snow_ice_percentage(time)float6431.3 27.28 27.47 ... 5.971 6.136
array([31.303981, 27.278802, 27.466634, 24.24368 , 8.633302, 12.746431, 12.460077, 5.774973, 0.34478 , 3.110733, 6.592238, 9.930623, 19.589281, 15.464897, 16.500069, 19.468927, 9.188948, 10.062376, 0.083632, 1.050132, 6.899765, 7.237989, 3.306192, 1.222734, 5.971161, 6.136174]) - mgrs:grid_square()<U2'PS'
array('PS', dtype='<U2') - s2:cloud_shadow_percentage(time)object0.123971 0.265839 ... 1.637513
array([0.123971, 0.265839, 1.426617, 1.504799, 4.112166, 2.610499, 4.382209, 3.633687, 0.766328, 0.140311, 1.309796, 2.989328, 0.735473, 0.90655, 0.556963, 0.533688, 3.479381, 3.173427, 0, 0.013745, 5.264171, 2.58029, 4.672229, 5.893223, 2.496737, 1.637513], dtype=object) - mgrs:utm_zone()int6432
array(32)
- s2:high_proba_clouds_percentage(time)float641.311 1.87 11.34 ... 8.549 9.308
array([ 1.310789, 1.869626, 11.341699, 11.836569, 41.895783, 42.292377, 37.297574, 37.018588, 78.666943, 79.059446, 45.465553, 45.84513 , 5.113235, 5.835326, 4.018103, 2.9164 , 23.718023, 24.355602, 27.910239, 27.134106, 16.794626, 17.557459, 31.651893, 31.285802, 8.549391, 9.307959]) - earthsearch:boa_offset_applied(time)boolTrue False True ... True True False
array([ True, False, True, False, True, False, False, True, True, False, True, False, True, False, False, True, True, False, False, True, True, False, False, True, True, False]) - s2:medium_proba_clouds_percentage(time)float641.2 2.886 5.046 ... 7.105 2.178
array([ 1.199887, 2.8858 , 5.045658, 3.289439, 15.328416, 5.294412, 6.934044, 18.3893 , 16.781795, 12.919861, 13.523927, 6.342981, 8.702251, 10.737985, 1.403487, 2.29385 , 11.548399, 3.597998, 69.088262, 68.578702, 9.756684, 2.711056, 4.114743, 12.67418 , 7.105023, 2.177899]) - s2:vegetation_percentage(time)float6440.25 40.69 44.01 ... 65.32 67.63
array([40.248781, 40.689981, 44.01297 , 45.765302, 24.552888, 27.177703, 30.178928, 28.761679, 1.449043, 2.117007, 22.656791, 23.482373, 46.2971 , 47.072074, 64.543813, 63.144523, 45.910549, 48.674297, 1.309192, 1.238064, 51.928717, 56.201601, 46.205747, 42.820308, 65.318006, 67.6287 ]) - s2:degraded_msi_data_percentage(time)object0.0112 0 0.0265 ... 0.1149 0.0127 0
array([0.0112, 0, 0.0265, 0, 0.1008, 0, 0, 0.2234, 0.1724, 0, 0.0443, 0, 0.0044, 0, 0, 0.0258, 0.0333, 0, 0, 0.0306, 0.0309, 0, 0, 0.1149, 0.0127, 0], dtype=object) - earthsearch:payload_id(time)<U74'roda-sentinel2/workflow-sentine...
array(['roda-sentinel2/workflow-sentinel2-to-stac/b84ebd104933e25116f9d2b158090bad', 'roda-sentinel2/workflow-sentinel2-to-stac/4565558a179b902dbd75791ba0821c05', 'roda-sentinel2/workflow-sentinel2-to-stac/9a9188736d6ffe2307874ac719c17178', 'roda-sentinel2/workflow-sentinel2-to-stac/fa9c01876d73bd7d39c8229cf48b4798', 'roda-sentinel2/workflow-sentinel2-to-stac/24be754211f90b660f6f3f82405e8c20', 'roda-sentinel2/workflow-sentinel2-to-stac/aa59d15d1c3bb88a838de30d9bca323a', 'roda-sentinel2/workflow-sentinel2-to-stac/c6b8893f8709d604a4501d85cae0ff1d', 'roda-sentinel2/workflow-sentinel2-to-stac/c2d5ccf5f168e5f3be4b6a420366f272', 'roda-sentinel2/workflow-sentinel2-to-stac/e7007d1cc5a002936cc6b3f3455f25d2', 'roda-sentinel2/workflow-sentinel2-to-stac/d8d561f2e1fddceff644293edac5a973', 'roda-sentinel2/workflow-sentinel2-to-stac/bfebbdc9c825add7648ced2d0f4795af', 'roda-sentinel2/workflow-sentinel2-to-stac/f806c3f35353dd870127f2df07aa0791', 'roda-sentinel2/workflow-sentinel2-to-stac/b156ec150b4550cf8b9d8456b9078007', 'roda-sentinel2/workflow-sentinel2-to-stac/2d850f82bac7b2a3b7d5daaade4ec1fc', 'roda-sentinel2/workflow-sentinel2-to-stac/8f4ef169c9800d925e7b78ed83b6243c', 'roda-sentinel2/workflow-sentinel2-to-stac/9a5972ef4da914562d2d2513790d027b', 'roda-sentinel2/workflow-sentinel2-to-stac/0318dd97ab3148f08142ef2bd1793c90', 'roda-sentinel2/workflow-sentinel2-to-stac/17239972afbdaaebb2408a7e0ebd5823', 'roda-sentinel2/workflow-sentinel2-to-stac/e0d4e2a19db22497be3d3ab319e5dd0a', 'roda-sentinel2/workflow-sentinel2-to-stac/d55883951e691ff8016328ad13ba5a80', 'roda-sentinel2/workflow-sentinel2-to-stac/ed7567a4bf3a0704cf8c35c32a534284', 'roda-sentinel2/workflow-sentinel2-to-stac/fde1a94a72e701b06ca16e3203c98fcc', 'roda-sentinel2/workflow-sentinel2-to-stac/98c0711e502f7af7cef9d582c2c5d4b1', 'roda-sentinel2/workflow-sentinel2-to-stac/9dc06d10fd3eef6b7f01c000dce4a184', 'roda-sentinel2/workflow-sentinel2-to-stac/39a848b9c950e88f80c1f5529b45d49a', 'roda-sentinel2/workflow-sentinel2-to-stac/c796a8ece534e903bb7d58398b39f859'], dtype='<U74') - gsd()int6410
array(10)
- proj:shape()object{10980}
array({10980}, dtype=object) - proj:transform()object{0, 600000, 10, 5200020, -10}
array({0, 600000, 10, 5200020, -10}, dtype=object) - epsg()int6432632
array(32632)
- timePandasIndex
PandasIndex(DatetimeIndex(['2021-05-28 10:17:59.882000', '2021-05-28 10:17:59.884000', '2021-05-31 10:27:56.286000', '2021-05-31 10:27:56.289000', '2021-06-02 10:18:00.103000', '2021-06-02 10:18:00.104000', '2021-06-05 10:27:56.280000', '2021-06-05 10:27:56.282000', '2021-06-07 10:18:00.323000', '2021-06-07 10:18:00.323000', '2021-06-10 10:27:56.565000', '2021-06-10 10:27:56.566000', '2021-06-12 10:17:59.823000', '2021-06-12 10:17:59.824000', '2021-06-15 10:27:55.829000', '2021-06-15 10:27:55.830000', '2021-06-17 10:18:00.191000', '2021-06-17 10:18:00.192000', '2021-06-20 10:27:56.235000', '2021-06-20 10:27:56.236000', '2021-06-22 10:18:00.204000', '2021-06-22 10:18:00.205000', '2021-06-25 10:27:56.865000', '2021-06-25 10:27:56.867000', '2021-06-27 10:18:00.077000', '2021-06-27 10:18:00.078000'], dtype='datetime64[ns]', name='time', freq=None)) - xPandasIndex
PandasIndex(Index([673310.0, 673320.0, 673330.0, 673340.0, 673350.0, 673360.0, 673370.0, 673380.0, 673390.0, 673400.0, ... 684670.0, 684680.0, 684690.0, 684700.0, 684710.0, 684720.0, 684730.0, 684740.0, 684750.0, 684760.0], dtype='float64', name='x', length=1146)) - yPandasIndex
PandasIndex(Index([5154890.0, 5154880.0, 5154870.0, 5154860.0, 5154850.0, 5154840.0, 5154830.0, 5154820.0, 5154810.0, 5154800.0, ... 5147850.0, 5147840.0, 5147830.0, 5147820.0, 5147810.0, 5147800.0, 5147790.0, 5147780.0, 5147770.0, 5147760.0], dtype='float64', name='y', length=714))
Quiz hint: look carefully at number of pixels of the loaded datacube!
The reducer could be again a single process, but when computing spectral indices like NDVI, NDSI etc. an arithmentical formula is used instead.
For instance, the NDVI formula can be expressed using a reduce_dimension process over the bands dimension:
def NDVI(data):
red = data.sel(band="red")
nir = data.sel(band="nir")
ndvi = (nir - red)/(nir + red)
return ndvi
ndvi_xr = NDVI(s2_cube)
ndvi_xr
<xarray.DataArray 'stackstac-b671372a6175b16d2e4eaf48f5606039' (time: 26,
y: 714, x: 1146)>
dask.array<truediv, shape=(26, 714, 1146), dtype=float64, chunksize=(1, 714, 1024), chunktype=numpy.ndarray>
Coordinates: (12/48)
* time (time) datetime64[ns] 2021-05-28...
id (time) <U24 'S2B_32TPS_20210528_...
* x (x) float64 6.733e+05 ... 6.848e+05
* y (y) float64 5.155e+06 ... 5.148e+06
s2:unclassified_percentage (time) float64 0.2507 ... 2.393
s2:product_type <U7 'S2MSI2A'
... ...
s2:degraded_msi_data_percentage (time) object 0.0112 0 ... 0.0127 0
earthsearch:payload_id (time) <U74 'roda-sentinel2/work...
gsd int64 10
proj:shape object {10980}
proj:transform object {0, 600000, 10, 5200020, ...
epsg int64 32632- time: 26
- y: 714
- x: 1146
- dask.array<chunksize=(1, 714, 1024), meta=np.ndarray>
Array Chunk Bytes 162.31 MiB 5.58 MiB Shape (26, 714, 1146) (1, 714, 1024) Dask graph 52 chunks in 10 graph layers Data type float64 numpy.ndarray - time(time)datetime64[ns]2021-05-28T10:17:59.882000 ... 2...
array(['2021-05-28T10:17:59.882000000', '2021-05-28T10:17:59.884000000', '2021-05-31T10:27:56.286000000', '2021-05-31T10:27:56.289000000', '2021-06-02T10:18:00.103000000', '2021-06-02T10:18:00.104000000', '2021-06-05T10:27:56.280000000', '2021-06-05T10:27:56.282000000', '2021-06-07T10:18:00.323000000', '2021-06-07T10:18:00.323000000', '2021-06-10T10:27:56.565000000', '2021-06-10T10:27:56.566000000', '2021-06-12T10:17:59.823000000', '2021-06-12T10:17:59.824000000', '2021-06-15T10:27:55.829000000', '2021-06-15T10:27:55.830000000', '2021-06-17T10:18:00.191000000', '2021-06-17T10:18:00.192000000', '2021-06-20T10:27:56.235000000', '2021-06-20T10:27:56.236000000', '2021-06-22T10:18:00.204000000', '2021-06-22T10:18:00.205000000', '2021-06-25T10:27:56.865000000', '2021-06-25T10:27:56.867000000', '2021-06-27T10:18:00.077000000', '2021-06-27T10:18:00.078000000'], dtype='datetime64[ns]') - id(time)<U24'S2B_32TPS_20210528_1_L2A' ... '...
array(['S2B_32TPS_20210528_1_L2A', 'S2B_32TPS_20210528_0_L2A', 'S2B_32TPS_20210531_1_L2A', 'S2B_32TPS_20210531_0_L2A', 'S2A_32TPS_20210602_1_L2A', 'S2A_32TPS_20210602_0_L2A', 'S2A_32TPS_20210605_0_L2A', 'S2A_32TPS_20210605_1_L2A', 'S2B_32TPS_20210607_1_L2A', 'S2B_32TPS_20210607_0_L2A', 'S2B_32TPS_20210610_1_L2A', 'S2B_32TPS_20210610_0_L2A', 'S2A_32TPS_20210612_1_L2A', 'S2A_32TPS_20210612_0_L2A', 'S2A_32TPS_20210615_0_L2A', 'S2A_32TPS_20210615_1_L2A', 'S2B_32TPS_20210617_1_L2A', 'S2B_32TPS_20210617_0_L2A', 'S2B_32TPS_20210620_0_L2A', 'S2B_32TPS_20210620_1_L2A', 'S2A_32TPS_20210622_1_L2A', 'S2A_32TPS_20210622_0_L2A', 'S2A_32TPS_20210625_0_L2A', 'S2A_32TPS_20210625_1_L2A', 'S2B_32TPS_20210627_1_L2A', 'S2B_32TPS_20210627_0_L2A'], dtype='<U24') - x(x)float646.733e+05 6.733e+05 ... 6.848e+05
array([673310., 673320., 673330., ..., 684740., 684750., 684760.])
- y(y)float645.155e+06 5.155e+06 ... 5.148e+06
array([5154890., 5154880., 5154870., ..., 5147780., 5147770., 5147760.])
- s2:unclassified_percentage(time)float640.2507 6.344 ... 0.3769 2.393
array([0.250706, 6.3441 , 0.515761, 3.860853, 1.493286, 5.219495, 4.114496, 1.357601, 0.809151, 1.985232, 2.379116, 4.723761, 2.041206, 4.504375, 2.619704, 0.152735, 0.65733 , 3.93955 , 0.770469, 0.292153, 0.427856, 3.074201, 3.535479, 0.935654, 0.376936, 2.392862]) - s2:product_type()<U7'S2MSI2A'
array('S2MSI2A', dtype='<U7') - s2:sequence(time)<U1'1' '0' '1' '0' ... '0' '1' '1' '0'
array(['1', '0', '1', '0', '1', '0', '0', '1', '1', '0', '1', '0', '1', '0', '0', '1', '1', '0', '0', '1', '1', '0', '0', '1', '1', '0'], dtype='<U1') - s2:generation_time(time)<U27'2023-02-27T08:23:12.000000Z' .....
array(['2023-02-27T08:23:12.000000Z', '2021-05-28T13:08:36.000000Z', '2023-03-01T00:24:41.000000Z', '2021-05-31T14:00:40.000000Z', '2023-03-13T04:04:11.000000Z', '2021-06-02T17:13:07.000000Z', '2021-06-05T15:23:29.000000Z', '2023-03-12T16:27:26.000000Z', '2023-03-14T07:14:12.000000Z', '2021-06-07T16:37:34.000000Z', '2023-03-21T10:24:57.000000Z', '2021-06-10T13:27:39.000000Z', '2023-03-16T11:37:02.000000Z', '2021-06-12T13:18:42.000000Z', '2021-06-15T13:16:59.000000Z', '2023-03-21T22:10:50.000000Z', '2023-03-21T22:17:28.000000Z', '2021-06-17T13:12:19.000000Z', '2021-06-20T14:05:52.000000Z', '2023-03-08T17:05:00.000000Z', '2023-01-29T15:30:31.000000Z', '2021-06-22T12:13:05.000000Z', '2021-06-25T13:25:02.000000Z', '2023-03-19T05:49:11.000000Z', '2023-03-18T23:38:59.000000Z', '2021-06-27T13:58:56.000000Z'], dtype='<U27') - s2:datatake_type()<U8'INS-NOBS'
array('INS-NOBS', dtype='<U8') - proj:epsg()int6432632
array(32632)
- mgrs:latitude_band()<U1'T'
array('T', dtype='<U1') - processing:software(time)object{'sentinel2-to-stac': '0.1.1'} ....
array([{'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}], dtype=object) - s2:processing_baseline(time)<U5'05.00' '03.00' ... '05.00' '03.00'
array(['05.00', '03.00', '05.00', '03.00', '05.00', '03.00', '03.00', '05.00', '05.00', '03.00', '05.00', '03.00', '05.00', '03.00', '03.00', '05.00', '05.00', '03.00', '03.00', '05.00', '05.00', '03.00', '03.00', '05.00', '05.00', '03.00'], dtype='<U5') - s2:datastrip_id(time)<U64'S2B_OPER_MSI_L2A_DS_S2RP_202302...
array(['S2B_OPER_MSI_L2A_DS_S2RP_20230227T082312_S20210528T100555_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20210528T130836_S20210528T100555_N03.00', 'S2B_OPER_MSI_L2A_DS_S2RP_20230301T002441_S20210531T102618_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS4_20210531T140040_S20210531T102618_N03.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20230313T040411_S20210602T101235_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210602T171307_S20210602T101235_N03.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210605T152329_S20210605T102737_N03.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20230312T162726_S20210605T102737_N05.00', 'S2B_OPER_MSI_L2A_DS_S2RP_20230314T071412_S20210607T101225_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20210607T163734_S20210607T101225_N03.00', 'S2B_OPER_MSI_L2A_DS_S2RP_20230321T102457_S20210610T102220_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20210610T132739_S20210610T102220_N03.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20230316T113702_S20210612T101329_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210612T131842_S20210612T101329_N03.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210615T131659_S20210615T102602_N03.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20230321T221050_S20210615T102602_N05.00', 'S2B_OPER_MSI_L2A_DS_S2RP_20230321T221728_S20210617T101301_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20210617T131219_S20210617T101301_N03.00', 'S2B_OPER_MSI_L2A_DS_VGS4_20210620T140552_S20210620T102722_N03.00', 'S2B_OPER_MSI_L2A_DS_S2RP_20230308T170500_S20210620T102722_N05.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20230129T153031_S20210622T101416_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS4_20210622T121305_S20210622T101416_N03.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210625T132502_S20210625T102545_N03.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20230319T054911_S20210625T102545_N05.00', 'S2B_OPER_MSI_L2A_DS_S2RP_20230318T233859_S20210627T101702_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS4_20210627T135856_S20210627T101702_N03.00'], dtype='<U64') - s2:product_uri(time)<U65'S2B_MSIL2A_20210528T100559_N050...
array(['S2B_MSIL2A_20210528T100559_N0500_R022_T32TPS_20230227T082312.SAFE', 'S2B_MSIL2A_20210528T100559_N0300_R022_T32TPS_20210528T130836.SAFE', 'S2B_MSIL2A_20210531T101559_N0500_R065_T32TPS_20230301T002441.SAFE', 'S2B_MSIL2A_20210531T101559_N0300_R065_T32TPS_20210531T140040.SAFE', 'S2A_MSIL2A_20210602T101031_N0500_R022_T32TPS_20230313T040411.SAFE', 'S2A_MSIL2A_20210602T101031_N0300_R022_T32TPS_20210602T171307.SAFE', 'S2A_MSIL2A_20210605T102021_N0300_R065_T32TPS_20210605T152329.SAFE', 'S2A_MSIL2A_20210605T102021_N0500_R065_T32TPS_20230312T162726.SAFE', 'S2B_MSIL2A_20210607T100559_N0500_R022_T32TPS_20230314T071412.SAFE', 'S2B_MSIL2A_20210607T100559_N0300_R022_T32TPS_20210607T163734.SAFE', 'S2B_MSIL2A_20210610T101559_N0500_R065_T32TPS_20230321T102457.SAFE', 'S2B_MSIL2A_20210610T101559_N0300_R065_T32TPS_20210610T132739.SAFE', 'S2A_MSIL2A_20210612T101031_N0500_R022_T32TPS_20230316T113702.SAFE', 'S2A_MSIL2A_20210612T101031_N0300_R022_T32TPS_20210612T131842.SAFE', 'S2A_MSIL2A_20210615T102021_N0300_R065_T32TPS_20210615T131659.SAFE', 'S2A_MSIL2A_20210615T102021_N0500_R065_T32TPS_20230321T221050.SAFE', 'S2B_MSIL2A_20210617T100559_N0500_R022_T32TPS_20230321T221728.SAFE', 'S2B_MSIL2A_20210617T100559_N0300_R022_T32TPS_20210617T131219.SAFE', 'S2B_MSIL2A_20210620T101559_N0300_R065_T32TPS_20210620T140552.SAFE', 'S2B_MSIL2A_20210620T101559_N0500_R065_T32TPS_20230308T170500.SAFE', 'S2A_MSIL2A_20210622T101031_N0500_R022_T32TPS_20230129T153031.SAFE', 'S2A_MSIL2A_20210622T101031_N0300_R022_T32TPS_20210622T121305.SAFE', 'S2A_MSIL2A_20210625T102021_N0300_R065_T32TPS_20210625T132502.SAFE', 'S2A_MSIL2A_20210625T102021_N0500_R065_T32TPS_20230319T054911.SAFE', 'S2B_MSIL2A_20210627T100559_N0500_R022_T32TPS_20230318T233859.SAFE', 'S2B_MSIL2A_20210627T100559_N0300_R022_T32TPS_20210627T135856.SAFE'], dtype='<U65') - eo:cloud_cover(time)float6412.65 14.92 16.5 ... 16.35 11.91
array([12.645555, 14.923827, 16.502707, 15.246907, 57.402623, 47.756503, 44.72053 , 56.068999, 96.218115, 92.314465, 62.109846, 55.815224, 21.750824, 24.577105, 5.444319, 5.673918, 35.295203, 27.991417, 97.720245, 96.397674, 26.698235, 20.468445, 36.142011, 44.22752 , 16.351688, 11.912692]) - instruments()<U3'msi'
array('msi', dtype='<U3') - s2:datatake_id(time)<U34'GS2B_20210528T100559_022070_N05...
array(['GS2B_20210528T100559_022070_N05.00', 'GS2B_20210528T100559_022070_N03.00', 'GS2B_20210531T101559_022113_N05.00', 'GS2B_20210531T101559_022113_N03.00', 'GS2A_20210602T101031_031050_N05.00', 'GS2A_20210602T101031_031050_N03.00', 'GS2A_20210605T102021_031093_N03.00', 'GS2A_20210605T102021_031093_N05.00', 'GS2B_20210607T100559_022213_N05.00', 'GS2B_20210607T100559_022213_N03.00', 'GS2B_20210610T101559_022256_N05.00', 'GS2B_20210610T101559_022256_N03.00', 'GS2A_20210612T101031_031193_N05.00', 'GS2A_20210612T101031_031193_N03.00', 'GS2A_20210615T102021_031236_N03.00', 'GS2A_20210615T102021_031236_N05.00', 'GS2B_20210617T100559_022356_N05.00', 'GS2B_20210617T100559_022356_N03.00', 'GS2B_20210620T101559_022399_N03.00', 'GS2B_20210620T101559_022399_N05.00', 'GS2A_20210622T101031_031336_N05.00', 'GS2A_20210622T101031_031336_N03.00', 'GS2A_20210625T102021_031379_N03.00', 'GS2A_20210625T102021_031379_N05.00', 'GS2B_20210627T100559_022499_N05.00', 'GS2B_20210627T100559_022499_N03.00'], dtype='<U34') - s2:nodata_pixel_percentage(time)object0 7e-06 11.874377 ... 0 7e-06
array([0, 7e-06, 11.874377, 11.646823, 0, 3e-06, 10.985302, 11.013988, 0, 0, 11.783398, 11.556501, 8.3e-05, 1.3e-05, 10.792472, 10.827903, 0, 1.7e-05, 11.396532, 11.627065, 0.000146, 1.7e-05, 10.879045, 10.92066, 0, 7e-06], dtype=object) - view:sun_azimuth(time)float64151.1 151.1 155.4 ... 146.8 146.8
array([151.07526534, 151.07388572, 155.39203616, 155.39059825, 150.16382152, 150.16244266, 154.53346131, 154.5349045 , 149.27864414, 149.2772675 , 153.70866815, 153.70724073, 148.4554036 , 148.45405463, 152.94501975, 152.94641223, 147.75003773, 147.74872101, 152.3076782 , 152.30905816, 147.19176534, 147.19046035, 151.8233914 , 151.82473308, 146.81010023, 146.80884882]) - earthsearch:s3_path(time)<U79's3://sentinel-cogs/sentinel-s2-...
array(['s3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/5/S2B_32TPS_20210528_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/5/S2B_32TPS_20210528_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/5/S2B_32TPS_20210531_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/5/S2B_32TPS_20210531_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210602_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210602_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210605_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210605_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210607_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210607_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210610_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210610_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210612_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210612_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210615_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210615_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210617_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210617_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210620_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210620_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210622_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210622_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210625_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2A_32TPS_20210625_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210627_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/6/S2B_32TPS_20210627_0_L2A'], dtype='<U79') - constellation()<U10'sentinel-2'
array('sentinel-2', dtype='<U10') - s2:granule_id(time)<U62'S2B_OPER_MSI_L2A_TL_S2RP_202302...
array(['S2B_OPER_MSI_L2A_TL_S2RP_20230227T082312_A022070_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20210528T130836_A022070_T32TPS_N03.00', 'S2B_OPER_MSI_L2A_TL_S2RP_20230301T002441_A022113_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS4_20210531T140040_A022113_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20230313T040411_A031050_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210602T171307_A031050_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210605T152329_A031093_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20230312T162726_A031093_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_S2RP_20230314T071412_A022213_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20210607T163734_A022213_T32TPS_N03.00', 'S2B_OPER_MSI_L2A_TL_S2RP_20230321T102457_A022256_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20210610T132739_A022256_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20230316T113702_A031193_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210612T131842_A031193_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210615T131659_A031236_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20230321T221050_A031236_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_S2RP_20230321T221728_A022356_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20210617T131219_A022356_T32TPS_N03.00', 'S2B_OPER_MSI_L2A_TL_VGS4_20210620T140552_A022399_T32TPS_N03.00', 'S2B_OPER_MSI_L2A_TL_S2RP_20230308T170500_A022399_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20230129T153031_A031336_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS4_20210622T121305_A031336_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210625T132502_A031379_T32TPS_N03.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20230319T054911_A031379_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_S2RP_20230318T233859_A022499_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS4_20210627T135856_A022499_T32TPS_N03.00'], dtype='<U62') - s2:reflectance_conversion_factor(time)float640.975 0.975 0.974 ... 0.9679 0.9679
array([0.97504789, 0.97504789, 0.97399904, 0.97399904, 0.97334113, 0.97334113, 0.97240967, 0.97240967, 0.97183347, 0.97183347, 0.97102521, 0.97102521, 0.97053088, 0.97053088, 0.96985123, 0.96985123, 0.969445 , 0.969445 , 0.96889809, 0.96889809, 0.96858002, 0.96858002, 0.96816954, 0.96816954, 0.96794362, 0.96794362]) - grid:code()<U10'MGRS-32TPS'
array('MGRS-32TPS', dtype='<U10') - updated(time)<U24'2023-07-19T01:31:45.362Z' ... '...
array(['2023-07-19T01:31:45.362Z', '2022-11-06T13:34:17.157Z', '2023-08-21T02:48:42.746Z', '2022-11-06T04:43:15.876Z', '2023-07-14T10:06:11.004Z', '2022-11-06T04:43:21.072Z', '2022-11-06T04:42:40.826Z', '2023-08-19T18:25:04.412Z', '2023-07-15T20:10:05.181Z', '2022-11-06T04:44:31.882Z', '2023-08-28T22:22:56.940Z', '2022-11-06T04:43:35.373Z', '2023-08-31T14:25:17.439Z', '2022-11-06T04:44:47.117Z', '2022-11-06T04:44:42.815Z', '2023-08-28T20:41:39.160Z', '2023-08-28T19:50:44.246Z', '2022-11-06T04:43:20.506Z', '2022-11-06T04:46:02.240Z', '2023-07-16T11:32:44.388Z', '2023-07-12T22:29:59.130Z', '2022-11-06T04:46:04.928Z', '2022-11-06T04:45:30.338Z', '2023-09-01T04:30:40.825Z', '2023-09-02T00:59:09.859Z', '2022-11-06T04:45:27.038Z'], dtype='<U24') - created(time)<U24'2023-07-19T01:31:45.362Z' ... '...
array(['2023-07-19T01:31:45.362Z', '2022-11-03T10:53:30.936Z', '2023-08-21T02:48:42.746Z', '2022-11-06T04:43:15.876Z', '2023-07-14T10:06:11.004Z', '2022-11-06T04:43:21.072Z', '2022-11-06T04:42:40.826Z', '2023-08-19T18:25:04.412Z', '2023-07-15T20:10:05.181Z', '2022-11-06T04:44:31.882Z', '2023-08-28T22:22:56.940Z', '2022-11-06T04:43:35.373Z', '2023-08-31T14:25:17.439Z', '2022-11-06T04:44:47.117Z', '2022-11-06T04:44:42.815Z', '2023-08-28T20:41:39.160Z', '2023-08-28T19:50:44.246Z', '2022-11-06T04:43:20.506Z', '2022-11-06T04:46:02.240Z', '2023-07-16T11:32:44.388Z', '2023-07-12T22:29:59.130Z', '2022-11-06T04:46:04.928Z', '2022-11-06T04:45:30.338Z', '2023-09-01T04:30:40.825Z', '2023-09-02T00:59:09.859Z', '2022-11-06T04:45:27.038Z'], dtype='<U24') - view:sun_elevation(time)float6462.68 62.67 63.84 ... 63.8 63.79
array([62.67508338, 62.67485352, 63.83829126, 63.83809076, 63.2776855 , 63.27744956, 64.36252912, 64.36273616, 63.71164562, 63.71140388, 64.71725324, 64.71704181, 63.97586903, 63.9756252 , 64.90059226, 64.90080535, 64.07538254, 64.07513775, 64.91877022, 64.91898737, 64.01302019, 64.0127717 , 64.77526927, 64.77548806, 63.79508809, 63.79484238]) - s2:thin_cirrus_percentage(time)float6410.13 10.17 ... 0.6973 0.4268
array([10.13488 , 10.168401, 0.115349, 0.120899, 0.178427, 0.169714, 0.488913, 0.661111, 0.769374, 0.335158, 3.120367, 3.627113, 7.935338, 8.003794, 0.022728, 0.463668, 0.028782, 0.037817, 0.721744, 0.684867, 0.146924, 0.19993 , 0.375375, 0.267541, 0.697274, 0.426833]) - platform(time)<U11'sentinel-2b' ... 'sentinel-2b'
array(['sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b'], dtype='<U11') - s2:water_percentage(time)object0.20837 0.197246 ... 0.337965
array([0.20837, 0.197246, 0.271758, 0.382846, 0.142355, 0.298234, 0.443727, 0.187946, 0.022512, 0.027246, 0.175192, 0.226552, 0.209528, 0.239395, 0.357188, 0.292813, 0.272607, 0.497115, 0, 0.000169, 0.233775, 0.335679, 0.405969, 0.275791, 0.281174, 0.337965], dtype=object) - s2:dark_features_percentage(time)float640.9701 0.6094 ... 0.6925 0.9415
array([9.701360e-01, 6.094340e-01, 2.015643e+00, 1.392244e+00, 3.438870e-01, 6.917930e-01, 8.403930e-01, 1.378928e+00, 1.390000e-03, 4.481100e-02, 1.882207e+00, 7.810120e-01, 8.046860e-01, 5.937070e-01, 7.546780e-01, 1.151714e+00, 5.513320e-01, 7.116930e-01, 2.617000e-03, 2.910000e-03, 4.811240e-01, 9.703290e-01, 1.271975e+00, 9.746800e-01, 6.925060e-01, 9.414770e-01]) - s2:not_vegetated_percentage(time)float6414.25 9.691 7.788 ... 8.512 9.013
array([14.248499, 9.690771, 7.787908, 7.603369, 3.319488, 3.499338, 2.859635, 2.836192, 0.388685, 0.260192, 2.894815, 2.051123, 8.5719 , 6.641897, 9.223264, 9.58168 , 4.644649, 4.95012 , 0.113843, 1.005147, 8.066355, 9.131461, 4.460402, 3.650089, 8.511797, 9.012622]) - s2:saturated_defective_pixel_percentage()int640
array(0)
- s2:snow_ice_percentage(time)float6431.3 27.28 27.47 ... 5.971 6.136
array([31.303981, 27.278802, 27.466634, 24.24368 , 8.633302, 12.746431, 12.460077, 5.774973, 0.34478 , 3.110733, 6.592238, 9.930623, 19.589281, 15.464897, 16.500069, 19.468927, 9.188948, 10.062376, 0.083632, 1.050132, 6.899765, 7.237989, 3.306192, 1.222734, 5.971161, 6.136174]) - mgrs:grid_square()<U2'PS'
array('PS', dtype='<U2') - s2:cloud_shadow_percentage(time)object0.123971 0.265839 ... 1.637513
array([0.123971, 0.265839, 1.426617, 1.504799, 4.112166, 2.610499, 4.382209, 3.633687, 0.766328, 0.140311, 1.309796, 2.989328, 0.735473, 0.90655, 0.556963, 0.533688, 3.479381, 3.173427, 0, 0.013745, 5.264171, 2.58029, 4.672229, 5.893223, 2.496737, 1.637513], dtype=object) - mgrs:utm_zone()int6432
array(32)
- s2:high_proba_clouds_percentage(time)float641.311 1.87 11.34 ... 8.549 9.308
array([ 1.310789, 1.869626, 11.341699, 11.836569, 41.895783, 42.292377, 37.297574, 37.018588, 78.666943, 79.059446, 45.465553, 45.84513 , 5.113235, 5.835326, 4.018103, 2.9164 , 23.718023, 24.355602, 27.910239, 27.134106, 16.794626, 17.557459, 31.651893, 31.285802, 8.549391, 9.307959]) - earthsearch:boa_offset_applied(time)boolTrue False True ... True True False
array([ True, False, True, False, True, False, False, True, True, False, True, False, True, False, False, True, True, False, False, True, True, False, False, True, True, False]) - s2:medium_proba_clouds_percentage(time)float641.2 2.886 5.046 ... 7.105 2.178
array([ 1.199887, 2.8858 , 5.045658, 3.289439, 15.328416, 5.294412, 6.934044, 18.3893 , 16.781795, 12.919861, 13.523927, 6.342981, 8.702251, 10.737985, 1.403487, 2.29385 , 11.548399, 3.597998, 69.088262, 68.578702, 9.756684, 2.711056, 4.114743, 12.67418 , 7.105023, 2.177899]) - s2:vegetation_percentage(time)float6440.25 40.69 44.01 ... 65.32 67.63
array([40.248781, 40.689981, 44.01297 , 45.765302, 24.552888, 27.177703, 30.178928, 28.761679, 1.449043, 2.117007, 22.656791, 23.482373, 46.2971 , 47.072074, 64.543813, 63.144523, 45.910549, 48.674297, 1.309192, 1.238064, 51.928717, 56.201601, 46.205747, 42.820308, 65.318006, 67.6287 ]) - s2:degraded_msi_data_percentage(time)object0.0112 0 0.0265 ... 0.1149 0.0127 0
array([0.0112, 0, 0.0265, 0, 0.1008, 0, 0, 0.2234, 0.1724, 0, 0.0443, 0, 0.0044, 0, 0, 0.0258, 0.0333, 0, 0, 0.0306, 0.0309, 0, 0, 0.1149, 0.0127, 0], dtype=object) - earthsearch:payload_id(time)<U74'roda-sentinel2/workflow-sentine...
array(['roda-sentinel2/workflow-sentinel2-to-stac/b84ebd104933e25116f9d2b158090bad', 'roda-sentinel2/workflow-sentinel2-to-stac/4565558a179b902dbd75791ba0821c05', 'roda-sentinel2/workflow-sentinel2-to-stac/9a9188736d6ffe2307874ac719c17178', 'roda-sentinel2/workflow-sentinel2-to-stac/fa9c01876d73bd7d39c8229cf48b4798', 'roda-sentinel2/workflow-sentinel2-to-stac/24be754211f90b660f6f3f82405e8c20', 'roda-sentinel2/workflow-sentinel2-to-stac/aa59d15d1c3bb88a838de30d9bca323a', 'roda-sentinel2/workflow-sentinel2-to-stac/c6b8893f8709d604a4501d85cae0ff1d', 'roda-sentinel2/workflow-sentinel2-to-stac/c2d5ccf5f168e5f3be4b6a420366f272', 'roda-sentinel2/workflow-sentinel2-to-stac/e7007d1cc5a002936cc6b3f3455f25d2', 'roda-sentinel2/workflow-sentinel2-to-stac/d8d561f2e1fddceff644293edac5a973', 'roda-sentinel2/workflow-sentinel2-to-stac/bfebbdc9c825add7648ced2d0f4795af', 'roda-sentinel2/workflow-sentinel2-to-stac/f806c3f35353dd870127f2df07aa0791', 'roda-sentinel2/workflow-sentinel2-to-stac/b156ec150b4550cf8b9d8456b9078007', 'roda-sentinel2/workflow-sentinel2-to-stac/2d850f82bac7b2a3b7d5daaade4ec1fc', 'roda-sentinel2/workflow-sentinel2-to-stac/8f4ef169c9800d925e7b78ed83b6243c', 'roda-sentinel2/workflow-sentinel2-to-stac/9a5972ef4da914562d2d2513790d027b', 'roda-sentinel2/workflow-sentinel2-to-stac/0318dd97ab3148f08142ef2bd1793c90', 'roda-sentinel2/workflow-sentinel2-to-stac/17239972afbdaaebb2408a7e0ebd5823', 'roda-sentinel2/workflow-sentinel2-to-stac/e0d4e2a19db22497be3d3ab319e5dd0a', 'roda-sentinel2/workflow-sentinel2-to-stac/d55883951e691ff8016328ad13ba5a80', 'roda-sentinel2/workflow-sentinel2-to-stac/ed7567a4bf3a0704cf8c35c32a534284', 'roda-sentinel2/workflow-sentinel2-to-stac/fde1a94a72e701b06ca16e3203c98fcc', 'roda-sentinel2/workflow-sentinel2-to-stac/98c0711e502f7af7cef9d582c2c5d4b1', 'roda-sentinel2/workflow-sentinel2-to-stac/9dc06d10fd3eef6b7f01c000dce4a184', 'roda-sentinel2/workflow-sentinel2-to-stac/39a848b9c950e88f80c1f5529b45d49a', 'roda-sentinel2/workflow-sentinel2-to-stac/c796a8ece534e903bb7d58398b39f859'], dtype='<U74') - gsd()int6410
array(10)
- proj:shape()object{10980}
array({10980}, dtype=object) - proj:transform()object{0, 600000, 10, 5200020, -10}
array({0, 600000, 10, 5200020, -10}, dtype=object) - epsg()int6432632
array(32632)
- timePandasIndex
PandasIndex(DatetimeIndex(['2021-05-28 10:17:59.882000', '2021-05-28 10:17:59.884000', '2021-05-31 10:27:56.286000', '2021-05-31 10:27:56.289000', '2021-06-02 10:18:00.103000', '2021-06-02 10:18:00.104000', '2021-06-05 10:27:56.280000', '2021-06-05 10:27:56.282000', '2021-06-07 10:18:00.323000', '2021-06-07 10:18:00.323000', '2021-06-10 10:27:56.565000', '2021-06-10 10:27:56.566000', '2021-06-12 10:17:59.823000', '2021-06-12 10:17:59.824000', '2021-06-15 10:27:55.829000', '2021-06-15 10:27:55.830000', '2021-06-17 10:18:00.191000', '2021-06-17 10:18:00.192000', '2021-06-20 10:27:56.235000', '2021-06-20 10:27:56.236000', '2021-06-22 10:18:00.204000', '2021-06-22 10:18:00.205000', '2021-06-25 10:27:56.865000', '2021-06-25 10:27:56.867000', '2021-06-27 10:18:00.077000', '2021-06-27 10:18:00.078000'], dtype='datetime64[ns]', name='time', freq=None)) - xPandasIndex
PandasIndex(Index([673310.0, 673320.0, 673330.0, 673340.0, 673350.0, 673360.0, 673370.0, 673380.0, 673390.0, 673400.0, ... 684670.0, 684680.0, 684690.0, 684700.0, 684710.0, 684720.0, 684730.0, 684740.0, 684750.0, 684760.0], dtype='float64', name='x', length=1146)) - yPandasIndex
PandasIndex(Index([5154890.0, 5154880.0, 5154870.0, 5154860.0, 5154850.0, 5154840.0, 5154830.0, 5154820.0, 5154810.0, 5154800.0, ... 5147850.0, 5147840.0, 5147830.0, 5147820.0, 5147810.0, 5147800.0, 5147790.0, 5147780.0, 5147770.0, 5147760.0], dtype='float64', name='y', length=714))
Visualize a sample NDVI result:
%%time
ndvi_xr.isel(time=0).plot.imshow(vmin=-1,vmax=1,cmap="Greens")
/srv/conda/envs/notebook/lib/python3.11/site-packages/dask/core.py:127: RuntimeWarning: invalid value encountered in divide
return func(*(_execute_task(a, cache) for a in args))
CPU times: user 650 ms, sys: 129 ms, total: 779 ms
Wall time: 4.33 s
<matplotlib.image.AxesImage at 0x7fc3b41bead0>
Additionally, it is possible to reduce both spatial dimensions of the datacube at the same time.
To do this, we need the reduce_spatial process.
This time we select a smaller area of interest, to reduce the amount of downloaded data:
# West, South, East, North
spatial_extent = [11.31369, 46.52167, 11.31906, 46.52425]
temporal_extent = ["2021-01-01T00:00:00Z","2021-12-30T00:00:00Z"]
bands = ["red","nir"]
URL = "https://earth-search.aws.element84.com/v1"
catalog = pystac_client.Client.open(URL)
s2_items = catalog.search(
bbox=spatial_extent,
datetime=temporal_extent,
collections=["sentinel-2-l2a"]
).item_collection()
s2_cube = stackstac.stack(s2_items,
bounds_latlon=spatial_extent,
assets=bands
)
s2_cube = s2_cube[s2_cube["eo:cloud_cover"] < 15]
/srv/conda/envs/notebook/lib/python3.11/site-packages/stackstac/prepare.py:408: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
Get rid of possible negative values#
s2_cube = xr.where(s2_cube>=0, s2_cube, 0)
Reduce dimension x and y with median values#
datacube_spatial_median = s2_cube.median(dim=["x", "y"])
Verify that the spatial dimensions were collapsed:
datacube_spatial_median
<xarray.DataArray 'stackstac-7710f08aba1196bc4f45cd73b40041b2' (time: 65,
band: 2)>
dask.array<nanmedian, shape=(65, 2), dtype=float64, chunksize=(1, 1), chunktype=numpy.ndarray>
Coordinates: (12/51)
* time (time) datetime64[ns] 2021-01-11...
id (time) <U24 'S2B_32TPS_20210111_...
* band (band) <U3 'red' 'nir'
s2:unclassified_percentage (time) object 0.001827 ... 5.000642
s2:product_type <U7 'S2MSI2A'
s2:sequence (time) <U1 '1' '3' '0' ... '0' '0'
... ...
proj:transform object {0, 600000, 10, 5200020, ...
title (band) <U20 'Red (band 4) - 10m'...
common_name (band) <U3 'red' 'nir'
center_wavelength (band) float64 0.665 0.842
full_width_half_max (band) float64 0.038 0.145
epsg int64 32632- time: 65
- band: 2
- dask.array<chunksize=(1, 1), meta=np.ndarray>
Array Chunk Bytes 1.02 kiB 8 B Shape (65, 2) (1, 1) Dask graph 130 chunks in 7 graph layers Data type float64 numpy.ndarray - time(time)datetime64[ns]2021-01-11T10:27:54.911000 ... 2...
array(['2021-01-11T10:27:54.911000000', '2021-02-15T10:27:54.551000000', '2021-02-15T10:27:54.552000000', '2021-02-17T10:17:57.062000000', '2021-02-17T10:17:57.062000000', '2021-02-20T10:27:53.334000000', '2021-02-20T10:27:53.335000000', '2021-02-22T10:17:58.873000000', '2021-02-22T10:17:58.873000000', '2021-02-25T10:27:55.240000000', '2021-02-25T10:27:55.241000000', '2021-03-02T10:27:54.438000000', '2021-03-02T10:27:54.439000000', '2021-03-07T10:27:55.298000000', '2021-03-07T10:27:55.300000000', '2021-03-22T10:27:54.580000000', '2021-03-24T10:17:57.711000000', '2021-03-24T10:17:57.712000000', '2021-04-01T10:27:53.659000000', '2021-04-08T10:17:56.404000000', '2021-04-08T10:17:56.404000000', '2021-04-23T10:17:55.224000000', '2021-04-23T10:17:55.227000000', '2021-05-26T10:27:56.205000000', '2021-05-28T10:17:59.882000000', '2021-05-28T10:17:59.884000000', '2021-06-15T10:27:55.829000000', '2021-06-15T10:27:55.830000000', '2021-06-27T10:18:00.078000000', '2021-07-12T10:18:02.331000000', '2021-07-20T10:27:57.564000000', '2021-08-14T10:27:58.017000000', '2021-08-26T10:17:57.394000000', '2021-09-10T10:17:59.484000000', '2021-09-10T10:17:59.484000000', '2021-09-18T10:27:51.968000000', '2021-09-25T10:17:57.577000000', '2021-09-25T10:17:57.578000000', '2021-09-28T10:27:54.449000000', '2021-09-28T10:27:54.449000000', '2021-09-30T10:18:03.604000000', '2021-09-30T10:18:03.604000000', '2021-10-08T10:27:56.160000000', '2021-10-08T10:27:56.160000000', '2021-10-10T10:18:04.559000000', '2021-10-10T10:18:04.560000000', '2021-10-13T10:28:00.872000000', '2021-10-15T10:18:00.588000000', '2021-10-18T10:27:56.971000000', '2021-10-18T10:27:56.973000000', '2021-10-20T10:18:04.648000000', '2021-10-23T10:28:00.726000000', '2021-10-23T10:28:00.728000000', '2021-10-28T10:27:56.929000000', '2021-11-07T10:27:55.834000000', '2021-11-12T10:27:57.394000000', '2021-11-24T10:17:55.331000000', '2021-12-12T10:27:49.590000000', '2021-12-12T10:27:49.591000000', '2021-12-12T10:27:56.297000000', '2021-12-17T10:27:49.270000000', '2021-12-17T10:27:49.271000000', '2021-12-19T10:17:59.960000000', '2021-12-19T10:17:59.962000000', '2021-12-22T10:27:56.749000000'], dtype='datetime64[ns]') - id(time)<U24'S2B_32TPS_20210111_1_L2A' ... '...
array(['S2B_32TPS_20210111_1_L2A', 'S2A_32TPS_20210215_3_L2A', 'S2A_32TPS_20210215_0_L2A', 'S2B_32TPS_20210217_1_L2A', 'S2B_32TPS_20210217_0_L2A', 'S2B_32TPS_20210220_1_L2A', 'S2B_32TPS_20210220_0_L2A', 'S2A_32TPS_20210222_1_L2A', 'S2A_32TPS_20210222_0_L2A', 'S2A_32TPS_20210225_0_L2A', 'S2A_32TPS_20210225_3_L2A', 'S2B_32TPS_20210302_0_L2A', 'S2B_32TPS_20210302_1_L2A', 'S2A_32TPS_20210307_0_L2A', 'S2A_32TPS_20210307_1_L2A', 'S2B_32TPS_20210322_0_L2A', 'S2A_32TPS_20210324_1_L2A', 'S2A_32TPS_20210324_0_L2A', 'S2B_32TPS_20210401_0_L2A', 'S2B_32TPS_20210408_2_L2A', 'S2B_32TPS_20210408_0_L2A', 'S2A_32TPS_20210423_2_L2A', 'S2A_32TPS_20210423_0_L2A', 'S2A_32TPS_20210526_0_L2A', 'S2B_32TPS_20210528_1_L2A', 'S2B_32TPS_20210528_0_L2A', 'S2A_32TPS_20210615_0_L2A', 'S2A_32TPS_20210615_1_L2A', 'S2B_32TPS_20210627_0_L2A', 'S2A_32TPS_20210712_0_L2A', 'S2B_32TPS_20210720_0_L2A', 'S2A_32TPS_20210814_0_L2A', 'S2B_32TPS_20210826_0_L2A', 'S2A_32TPS_20210910_1_L2A', 'S2A_32TPS_20210910_0_L2A', 'S2B_32TPS_20210918_0_L2A', 'S2B_32TPS_20210925_1_L2A', 'S2B_32TPS_20210925_0_L2A', 'S2B_32TPS_20210928_1_L2A', 'S2B_32TPS_20210928_0_L2A', 'S2A_32TPS_20210930_1_L2A', 'S2A_32TPS_20210930_0_L2A', 'S2B_32TPS_20211008_1_L2A', 'S2B_32TPS_20211008_0_L2A', 'S2A_32TPS_20211010_1_L2A', 'S2A_32TPS_20211010_0_L2A', 'S2A_32TPS_20211013_2_L2A', 'S2B_32TPS_20211015_0_L2A', 'S2B_32TPS_20211018_1_L2A', 'S2B_32TPS_20211018_0_L2A', 'S2A_32TPS_20211020_0_L2A', 'S2A_32TPS_20211023_0_L2A', 'S2A_32TPS_20211023_3_L2A', 'S2B_32TPS_20211028_0_L2A', 'S2B_32TPS_20211107_0_L2A', 'S2A_32TPS_20211112_1_L2A', 'S2B_32TPS_20211124_0_L2A', 'S2A_32TPS_20211212_2_L2A', 'S2A_32TPS_20211212_1_L2A', 'S2A_32TPS_20211212_0_L2A', 'S2B_32TPS_20211217_0_L2A', 'S2B_32TPS_20211217_1_L2A', 'S2A_32TPS_20211219_1_L2A', 'S2A_32TPS_20211219_0_L2A', 'S2A_32TPS_20211222_0_L2A'], dtype='<U24') - band(band)<U3'red' 'nir'
array(['red', 'nir'], dtype='<U3')
- s2:unclassified_percentage(time)object0.001827 0.129823 ... 5.000642
array([0.001827, 0.129823, 5.903982, 0.004486, 4.831852, 0.174702, 6.857514, 0.000936, 6.597646, 5.502601, 0.005526, 5.38119, 0.003286, 5.514648, 0.013578, 5.421813, 0.000571, 5.279551, 5.857176, 0.227375, 5.52128, 0.308397, 4.807, 4.171408, 0.250706, 6.3441, 2.619704, 0.152735, 2.392862, 3.476528, 4.482188, 2.17731, 3.061006, 0.708835, 2.723365, 4.260587, 0.124747, 2.356505, 0.831079, 4.109764, 0.574301, 4.626107, 2.024649, 5.294078, 0.072926, 4.682079, 0.765953, 4.617824, 0.000803, 6.356327, 4.773093, 7.133023, 0.034025, 8.035865, 5.403754, 5.800857, 5.32987, 0.007234, 5.996048, 6.082753, 4.698034, 0.001508, 0.001105, 4.838076, 5.000642], dtype=object) - s2:product_type()<U7'S2MSI2A'
array('S2MSI2A', dtype='<U7') - s2:sequence(time)<U1'1' '3' '0' '1' ... '1' '1' '0' '0'
array(['1', '3', '0', '1', '0', '1', '0', '1', '0', '0', '3', '0', '1', '0', '1', '0', '1', '0', '0', '2', '0', '2', '0', '0', '1', '0', '0', '1', '0', '0', '0', '0', '0', '1', '0', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '2', '0', '1', '0', '0', '0', '3', '0', '0', '1', '0', '2', '1', '0', '0', '1', '1', '0', '0'], dtype='<U1') - s2:generation_time(time)<U27'2023-05-31T05:43:03.000000Z' .....
array(['2023-05-31T05:43:03.000000Z', '2023-06-09T01:27:37.000000Z', '2021-02-15T13:11:02.000000Z', '2023-05-21T16:56:36.000000Z', '2021-02-17T12:09:10.000000Z', '2023-05-19T05:06:36.000000Z', '2021-02-20T13:09:38.000000Z', '2023-05-21T12:51:13.000000Z', '2021-02-22T12:01:41.000000Z', '2021-02-25T13:12:30.000000Z', '2023-06-04T11:19:35.000000Z', '2021-03-02T13:08:05.000000Z', '2023-06-06T02:35:53.000000Z', '2021-03-07T13:16:14.000000Z', '2023-05-25T12:31:49.000000Z', '2021-03-22T13:56:15.000000Z', '2023-06-08T01:43:47.000000Z', '2021-03-24T14:34:56.000000Z', '2021-04-01T14:19:13.000000Z', '2023-05-13T12:44:55.000000Z', '2021-04-08T13:26:17.000000Z', '2023-05-12T19:45:03.000000Z', '2021-04-23T12:55:30.000000Z', '2021-05-26T13:34:25.000000Z', '2023-02-27T08:23:12.000000Z', '2021-05-28T13:08:36.000000Z', '2021-06-15T13:16:59.000000Z', '2023-03-21T22:10:50.000000Z', '2021-06-27T13:58:56.000000Z', '2021-07-12T13:22:19.000000Z', '2021-07-20T13:22:07.000000Z', '2021-08-14T13:23:26.000000Z', '2021-08-26T13:11:06.000000Z', '2023-01-16T19:33:32.000000Z', '2021-09-10T13:19:06.000000Z', '2021-09-18T13:51:51.000000Z', '2023-01-21T20:42:23.000000Z', '2021-09-25T13:47:01.000000Z', '2023-01-25T15:55:19.000000Z', '2021-09-28T13:49:45.000000Z', '2023-01-15T19:24:58.000000Z', '2021-09-30T11:58:00.000000Z', '2023-01-12T12:46:37.000000Z', '2021-10-08T13:24:26.000000Z', '2023-01-13T16:47:44.000000Z', '2021-10-10T13:19:25.000000Z', '2023-01-15T05:24:55.000000Z', '2021-10-15T13:16:08.000000Z', '2023-01-11T00:14:02.000000Z', '2021-10-18T13:18:32.000000Z', '2021-10-20T12:04:20.000000Z', '2021-10-23T13:24:00.000000Z', '2023-01-12T12:45:05.000000Z', '2021-10-28T12:19:42.000000Z', '2021-11-07T12:32:50.000000Z', '2021-11-12T13:19:01.000000Z', '2021-11-24T12:26:23.000000Z', '2022-12-24T05:25:49.000000Z', '2021-12-12T11:50:21.000000Z', '2021-12-12T13:17:17.000000Z', '2021-12-17T12:22:56.000000Z', '2022-12-28T16:01:51.000000Z', '2022-12-27T16:30:48.000000Z', '2021-12-19T12:06:54.000000Z', '2021-12-22T13:16:13.000000Z'], dtype='<U27') - s2:datatake_type()<U8'INS-NOBS'
array('INS-NOBS', dtype='<U8') - proj:epsg()int6432632
array(32632)
- mgrs:latitude_band()<U1'T'
array('T', dtype='<U1') - processing:software(time)object{'sentinel2-to-stac': '0.1.1'} ....
array([{'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}], dtype=object) - s2:processing_baseline(time)<U5'05.00' '05.00' ... '03.01' '03.01'
array(['05.00', '05.00', '02.14', '05.00', '02.14', '05.00', '02.14', '05.00', '02.14', '02.14', '05.00', '02.14', '05.00', '02.14', '05.00', '02.14', '05.00', '02.14', '03.00', '05.00', '03.00', '05.00', '03.00', '03.00', '05.00', '03.00', '03.00', '05.00', '03.00', '03.01', '03.01', '03.01', '03.01', '05.00', '03.01', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '03.01', '03.01', '05.00', '03.01', '03.01', '03.01', '03.01', '05.00', '03.01', '03.01', '03.01', '05.00', '05.00', '03.01', '03.01'], dtype='<U5') - s2:datastrip_id(time)<U64'S2B_OPER_MSI_L2A_DS_S2RP_202305...
array(['S2B_OPER_MSI_L2A_DS_S2RP_20230531T054303_S20210111T102336_N05.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20230609T012737_S20210215T102741_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210215T131102_S20210215T102741_N02.14', 'S2B_OPER_MSI_L2A_DS_S2RP_20230521T165636_S20210217T101023_N05.00', 'S2B_OPER_MSI_L2A_DS_EPAE_20210217T120910_S20210217T101023_N02.14', 'S2B_OPER_MSI_L2A_DS_S2RP_20230519T050636_S20210220T102440_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20210220T130938_S20210220T102440_N02.14', 'S2A_OPER_MSI_L2A_DS_S2RP_20230521T125113_S20210222T101415_N05.00', 'S2A_OPER_MSI_L2A_DS_EPAE_20210222T120141_S20210222T101415_N02.14', 'S2A_OPER_MSI_L2A_DS_VGS2_20210225T131230_S20210225T102740_N02.14', 'S2A_OPER_MSI_L2A_DS_S2RP_20230604T111935_S20210225T102740_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20210302T130805_S20210302T101835_N02.14', 'S2B_OPER_MSI_L2A_DS_S2RP_20230606T023553_S20210302T101835_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210307T131614_S20210307T102135_N02.14', 'S2A_OPER_MSI_L2A_DS_S2RP_20230525T123149_S20210307T102135_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS4_20210322T135615_S20210322T102636_N02.14', 'S2A_OPER_MSI_L2A_DS_S2RP_20230608T014347_S20210324T101606_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210324T143456_S20210324T101606_N02.14', 'S2B_OPER_MSI_L2A_DS_VGS4_20210401T141913_S20210401T102326_N03.00', 'S2B_OPER_MSI_L2A_DS_S2RP_20230513T124455_S20210408T101148_N05.00', ... 'S2A_OPER_MSI_L2A_DS_S2RP_20230115T052455_S20211013T102746_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20211015T131608_S20211015T101504_N03.01', 'S2B_OPER_MSI_L2A_DS_S2RP_20230111T001402_S20211018T102105_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20211018T131832_S20211018T102105_N03.01', 'S2A_OPER_MSI_L2A_DS_VGS4_20211020T120420_S20211020T101421_N03.01', 'S2A_OPER_MSI_L2A_DS_VGS2_20211023T132400_S20211023T102744_N03.01', 'S2A_OPER_MSI_L2A_DS_S2RP_20230112T124505_S20211023T102744_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS4_20211028T121942_S20211028T102038_N03.01', 'S2B_OPER_MSI_L2A_DS_VGS4_20211107T123250_S20211107T102157_N03.01', 'S2A_OPER_MSI_L2A_DS_VGS2_20211112T131901_S20211112T102742_N03.01', 'S2B_OPER_MSI_L2A_DS_VGS4_20211124T122623_S20211124T101240_N03.01', 'S2A_OPER_MSI_L2A_DS_S2RP_20221224T052549_S20211212T102427_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS4_20211212T115021_S20211212T102427_N03.01', 'S2A_OPER_MSI_L2A_DS_VGS2_20211212T131717_S20211212T102745_N03.01', 'S2B_OPER_MSI_L2A_DS_VGS4_20211217T122256_S20211217T102332_N03.01', 'S2B_OPER_MSI_L2A_DS_S2RP_20221228T160151_S20211217T102332_N05.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20221227T163048_S20211219T101431_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS4_20211219T120654_S20211219T101431_N03.01', 'S2A_OPER_MSI_L2A_DS_VGS2_20211222T131613_S20211222T102744_N03.01'], dtype='<U64') - s2:product_uri(time)<U65'S2B_MSIL2A_20210111T102309_N050...
array(['S2B_MSIL2A_20210111T102309_N0500_R065_T32TPS_20230531T054303.SAFE', 'S2A_MSIL2A_20210215T102121_N0500_R065_T32TPS_20230609T012737.SAFE', 'S2A_MSIL2A_20210215T102121_N0214_R065_T32TPS_20210215T131102.SAFE', 'S2B_MSIL2A_20210217T101029_N0500_R022_T32TPS_20230521T165636.SAFE', 'S2B_MSIL2A_20210217T101029_N0214_R022_T32TPS_20210217T120910.SAFE', 'S2B_MSIL2A_20210220T101939_N0500_R065_T32TPS_20230519T050636.SAFE', 'S2B_MSIL2A_20210220T101939_N0214_R065_T32TPS_20210220T130938.SAFE', 'S2A_MSIL2A_20210222T101021_N0500_R022_T32TPS_20230521T125113.SAFE', 'S2A_MSIL2A_20210222T101021_N0214_R022_T32TPS_20210222T120141.SAFE', 'S2A_MSIL2A_20210225T102021_N0214_R065_T32TPS_20210225T131230.SAFE', 'S2A_MSIL2A_20210225T102021_N0500_R065_T32TPS_20230604T111935.SAFE', 'S2B_MSIL2A_20210302T101839_N0214_R065_T32TPS_20210302T130805.SAFE', 'S2B_MSIL2A_20210302T101839_N0500_R065_T32TPS_20230606T023553.SAFE', 'S2A_MSIL2A_20210307T102021_N0214_R065_T32TPS_20210307T131614.SAFE', 'S2A_MSIL2A_20210307T102021_N0500_R065_T32TPS_20230525T123149.SAFE', 'S2B_MSIL2A_20210322T101649_N0214_R065_T32TPS_20210322T135615.SAFE', 'S2A_MSIL2A_20210324T101021_N0500_R022_T32TPS_20230608T014347.SAFE', 'S2A_MSIL2A_20210324T101021_N0214_R022_T32TPS_20210324T143456.SAFE', 'S2B_MSIL2A_20210401T101559_N0300_R065_T32TPS_20210401T141913.SAFE', 'S2B_MSIL2A_20210408T100549_N0500_R022_T32TPS_20230513T124455.SAFE', ... 'S2A_MSIL2A_20211013T101951_N0500_R065_T32TPS_20230115T052455.SAFE', 'S2B_MSIL2A_20211015T101029_N0301_R022_T32TPS_20211015T131608.SAFE', 'S2B_MSIL2A_20211018T101939_N0500_R065_T32TPS_20230111T001402.SAFE', 'S2B_MSIL2A_20211018T101939_N0301_R065_T32TPS_20211018T131832.SAFE', 'S2A_MSIL2A_20211020T101041_N0301_R022_T32TPS_20211020T120420.SAFE', 'S2A_MSIL2A_20211023T102101_N0301_R065_T32TPS_20211023T132400.SAFE', 'S2A_MSIL2A_20211023T102101_N0500_R065_T32TPS_20230112T124505.SAFE', 'S2B_MSIL2A_20211028T102039_N0301_R065_T32TPS_20211028T121942.SAFE', 'S2B_MSIL2A_20211107T102129_N0301_R065_T32TPS_20211107T123250.SAFE', 'S2A_MSIL2A_20211112T102251_N0301_R065_T32TPS_20211112T131901.SAFE', 'S2B_MSIL2A_20211124T101239_N0301_R022_T32TPS_20211124T122623.SAFE', 'S2A_MSIL2A_20211212T102431_N0500_R065_T32TPS_20221224T052549.SAFE', 'S2A_MSIL2A_20211212T102431_N0301_R065_T32TPS_20211212T115021.SAFE', 'S2A_MSIL2A_20211212T102431_N0301_R065_T32TPS_20211212T131717.SAFE', 'S2B_MSIL2A_20211217T102329_N0301_R065_T32TPS_20211217T122256.SAFE', 'S2B_MSIL2A_20211217T102329_N0500_R065_T32TPS_20221228T160151.SAFE', 'S2A_MSIL2A_20211219T101431_N0500_R022_T32TPS_20221227T163048.SAFE', 'S2A_MSIL2A_20211219T101431_N0301_R022_T32TPS_20211219T120654.SAFE', 'S2A_MSIL2A_20211222T102441_N0301_R065_T32TPS_20211222T131613.SAFE'], dtype='<U65') - eo:cloud_cover(time)float642.857 9.315 11.37 ... 2.215 7.499
array([2.8569030e+00, 9.3151780e+00, 1.1371362e+01, 1.9023200e-01, 8.3656320e+00, 6.6702850e+00, 4.5112540e+00, 5.0381000e-02, 3.9241370e+00, 6.8572500e+00, 5.7320000e-02, 7.3764900e+00, 9.8729000e-02, 7.1364790e+00, 1.2444880e+00, 1.2101794e+01, 1.9612000e-02, 1.6021150e+00, 1.2815130e+00, 1.6437540e+00, 1.4011749e+01, 4.9645330e+00, 6.1402800e+00, 1.1467405e+01, 1.2645555e+01, 1.4923827e+01, 5.4443190e+00, 5.6739180e+00, 1.1912692e+01, 1.1819513e+01, 1.3046401e+01, 2.9289930e+00, 7.6342870e+00, 1.3073093e+01, 9.3538160e+00, 1.1764671e+01, 1.2179405e+01, 1.2935878e+01, 1.2853540e+01, 1.1459897e+01, 1.2891503e+01, 9.9084770e+00, 1.4478517e+01, 1.3405893e+01, 1.4375870e+00, 4.2042680e+00, 1.4188600e+01, 1.0860672e+01, 8.0150000e-03, 1.1780210e+00, 1.3292886e+01, 1.6803640e+00, 7.3226000e-01, 1.3601600e+00, 1.2108641e+01, 1.0056364e+01, 1.8670890e+00, 7.6005000e-02, 3.9374070e+00, 3.9650860e+00, 5.3958520e+00, 3.1637000e-02, 2.6410000e-03, 2.2150660e+00, 7.4991000e+00]) - instruments()<U3'msi'
array('msi', dtype='<U3') - s2:datatake_id(time)<U34'GS2B_20210111T102309_020111_N05...
array(['GS2B_20210111T102309_020111_N05.00', 'GS2A_20210215T102121_029520_N05.00', 'GS2A_20210215T102121_029520_N02.14', 'GS2B_20210217T101029_020640_N05.00', 'GS2B_20210217T101029_020640_N02.14', 'GS2B_20210220T101939_020683_N05.00', 'GS2B_20210220T101939_020683_N02.14', 'GS2A_20210222T101021_029620_N05.00', 'GS2A_20210222T101021_029620_N02.14', 'GS2A_20210225T102021_029663_N02.14', 'GS2A_20210225T102021_029663_N05.00', 'GS2B_20210302T101839_020826_N02.14', 'GS2B_20210302T101839_020826_N05.00', 'GS2A_20210307T102021_029806_N02.14', 'GS2A_20210307T102021_029806_N05.00', 'GS2B_20210322T101649_021112_N02.14', 'GS2A_20210324T101021_030049_N05.00', 'GS2A_20210324T101021_030049_N02.14', 'GS2B_20210401T101559_021255_N03.00', 'GS2B_20210408T100549_021355_N05.00', ... 'GS2A_20211010T100941_032909_N03.01', 'GS2A_20211013T101951_032952_N05.00', 'GS2B_20211015T101029_024072_N03.01', 'GS2B_20211018T101939_024115_N05.00', 'GS2B_20211018T101939_024115_N03.01', 'GS2A_20211020T101041_033052_N03.01', 'GS2A_20211023T102101_033095_N03.01', 'GS2A_20211023T102101_033095_N05.00', 'GS2B_20211028T102039_024258_N03.01', 'GS2B_20211107T102129_024401_N03.01', 'GS2A_20211112T102251_033381_N03.01', 'GS2B_20211124T101239_024644_N03.01', 'GS2A_20211212T102431_033810_N05.00', 'GS2A_20211212T102431_033810_N03.01', 'GS2A_20211212T102431_033810_N03.01', 'GS2B_20211217T102329_024973_N03.01', 'GS2B_20211217T102329_024973_N05.00', 'GS2A_20211219T101431_033910_N05.00', 'GS2A_20211219T101431_033910_N03.01', 'GS2A_20211222T102441_033953_N03.01'], dtype='<U34') - s2:nodata_pixel_percentage(time)object11.356236 12.297063 ... 12.545553
array([11.356236, 12.297063, 12.25481, 6.6e-05, 0.000202, 11.384106, 11.153702, 1e-05, 0.000143, 11.5767, 11.612403, 11.321751, 11.543947, 11.545821, 11.591096, 11.302146, 1.3e-05, 0.000182, 11.211243, 2.3e-05, 7.3e-05, 0.005421, 0.002943, 11.126715, 0, 7e-06, 10.792472, 10.827903, 7e-06, 3e-06, 11.535111, 11.207345, 3e-05, 0.00077, 0.000232, 10.601236, 0, 1e-05, 11.48045, 11.250049, 3e-06, 0.000166, 11.772808, 11.547185, 7e-06, 0.000292, 11.806361, 0.000196, 12.032601, 11.805047, 8.3e-05, 11.74587, 11.77223, 12.020504, 11.874768, 11.338881, 0.000448, 66.107917, 66.058993, 27.93659, 10.693993, 10.921072, 4.3e-05, 0.000471, 12.545553], dtype=object) - view:sun_azimuth(time)float64166.1 162.5 162.5 ... 166.6 168.7
array([166.11122138, 162.54877766, 162.54796386, 159.67290702, 159.67208988, 162.22097915, 162.2201451 , 159.31353071, 159.31270178, 161.95804731, 161.95888786, 161.72957356, 161.73043893, 161.54885084, 161.54974115, 161.1584357 , 157.7958327 , 157.79488336, 160.95467327, 157.11384099, 157.11279214, 156.0632289 , 156.06206564, 156.23770514, 151.07526534, 151.07388572, 152.94501975, 152.94641223, 146.80884882, 146.89062175, 152.21378548, 156.8151133 , 155.88046704, 159.86303814, 159.86238306, 165.21262305, 163.40604336, 163.40545498, 167.26091541, 167.26033544, 164.4675583 , 164.46699671, 168.95169412, 168.95114043, 166.23954491, 166.23900125, 169.66922933, 166.94828604, 170.24027851, 170.23974912, 167.58818255, 170.7407418 , 170.74126393, 171.09431652, 171.49581873, 171.5431442 , 168.70337048, 169.79854654, 169.79808633, 169.79808633, 169.2357253 , 169.23619963, 166.6458662 , 166.64539797, 168.6820382 ]) - earthsearch:s3_path(time)<U80's3://sentinel-cogs/sentinel-s2-...
array(['s3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/1/S2B_32TPS_20210111_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2A_32TPS_20210215_3_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2A_32TPS_20210215_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2B_32TPS_20210217_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2B_32TPS_20210217_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2B_32TPS_20210220_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2B_32TPS_20210220_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2A_32TPS_20210222_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2A_32TPS_20210222_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2A_32TPS_20210225_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2A_32TPS_20210225_3_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/3/S2B_32TPS_20210302_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/3/S2B_32TPS_20210302_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/3/S2A_32TPS_20210307_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/3/S2A_32TPS_20210307_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/3/S2B_32TPS_20210322_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/3/S2A_32TPS_20210324_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/3/S2A_32TPS_20210324_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/4/S2B_32TPS_20210401_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/4/S2B_32TPS_20210408_2_L2A', ... 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2A_32TPS_20211013_2_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2B_32TPS_20211015_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2B_32TPS_20211018_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2B_32TPS_20211018_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2A_32TPS_20211020_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2A_32TPS_20211023_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2A_32TPS_20211023_3_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2B_32TPS_20211028_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/11/S2B_32TPS_20211107_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/11/S2A_32TPS_20211112_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/11/S2B_32TPS_20211124_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211212_2_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211212_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211212_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2B_32TPS_20211217_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2B_32TPS_20211217_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211219_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211219_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211222_0_L2A'], dtype='<U80') - constellation()<U10'sentinel-2'
array('sentinel-2', dtype='<U10') - s2:granule_id(time)<U62'S2B_OPER_MSI_L2A_TL_S2RP_202305...
array(['S2B_OPER_MSI_L2A_TL_S2RP_20230531T054303_A020111_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20230609T012737_A029520_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210215T131102_A029520_T32TPS_N02.14', 'S2B_OPER_MSI_L2A_TL_S2RP_20230521T165636_A020640_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_EPAE_20210217T120910_A020640_T32TPS_N02.14', 'S2B_OPER_MSI_L2A_TL_S2RP_20230519T050636_A020683_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20210220T130938_A020683_T32TPS_N02.14', 'S2A_OPER_MSI_L2A_TL_S2RP_20230521T125113_A029620_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_EPAE_20210222T120141_A029620_T32TPS_N02.14', 'S2A_OPER_MSI_L2A_TL_VGS2_20210225T131230_A029663_T32TPS_N02.14', 'S2A_OPER_MSI_L2A_TL_S2RP_20230604T111935_A029663_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20210302T130805_A020826_T32TPS_N02.14', 'S2B_OPER_MSI_L2A_TL_S2RP_20230606T023553_A020826_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210307T131614_A029806_T32TPS_N02.14', 'S2A_OPER_MSI_L2A_TL_S2RP_20230525T123149_A029806_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS4_20210322T135615_A021112_T32TPS_N02.14', 'S2A_OPER_MSI_L2A_TL_S2RP_20230608T014347_A030049_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210324T143456_A030049_T32TPS_N02.14', 'S2B_OPER_MSI_L2A_TL_VGS4_20210401T141913_A021255_T32TPS_N03.00', 'S2B_OPER_MSI_L2A_TL_S2RP_20230513T124455_A021355_T32TPS_N05.00', ... 'S2A_OPER_MSI_L2A_TL_S2RP_20230115T052455_A032952_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20211015T131608_A024072_T32TPS_N03.01', 'S2B_OPER_MSI_L2A_TL_S2RP_20230111T001402_A024115_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20211018T131832_A024115_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_VGS4_20211020T120420_A033052_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_VGS2_20211023T132400_A033095_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_S2RP_20230112T124505_A033095_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS4_20211028T121942_A024258_T32TPS_N03.01', 'S2B_OPER_MSI_L2A_TL_VGS4_20211107T123250_A024401_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_VGS2_20211112T131901_A033381_T32TPS_N03.01', 'S2B_OPER_MSI_L2A_TL_VGS4_20211124T122623_A024644_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_S2RP_20221224T052549_A033810_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS4_20211212T115021_A033810_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_VGS2_20211212T131717_A033810_T32TPS_N03.01', 'S2B_OPER_MSI_L2A_TL_VGS4_20211217T122256_A024973_T32TPS_N03.01', 'S2B_OPER_MSI_L2A_TL_S2RP_20221228T160151_A024973_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20221227T163048_A033910_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS4_20211219T120654_A033910_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_VGS2_20211222T131613_A033953_T32TPS_N03.01'], dtype='<U62') - s2:reflectance_conversion_factor(time)float641.034 1.027 1.027 ... 1.033 1.033
array([1.03421211, 1.02653718, 1.02653718, 1.02577207, 1.02577207, 1.02456125, 1.02456125, 1.0237225 , 1.0237225 , 1.02240605, 1.02240605, 1.02009057, 1.02009057, 1.01763162, 1.01763162, 1.00960406, 1.0084828 , 1.0084828 , 1.00391722, 0.99989016, 0.99989016, 0.99141059, 0.99141059, 0.97577919, 0.97504789, 0.97504789, 0.96985123, 0.96985123, 0.96794362, 0.96743179, 0.96802315, 0.97358286, 0.97804701, 0.98495124, 0.98495124, 0.98910253, 0.9929235 , 0.9929235 , 0.99460618, 0.99460618, 0.99573409, 0.99573409, 1.00031599, 1.00031599, 1.00146456, 1.00146456, 1.00319682, 1.00434323, 1.0060657 , 1.0060657 , 1.00720174, 1.00890228, 1.00890228, 1.01168368, 1.01699809, 1.01948942, 1.02486761, 1.03092533, 1.03092533, 1.03092533, 1.03209956, 1.03209956, 1.03250105, 1.03250105, 1.0330328 ]) - grid:code()<U10'MGRS-32TPS'
array('MGRS-32TPS', dtype='<U10') - updated(time)<U24'2023-08-30T09:53:24.991Z' ... '...
array(['2023-08-30T09:53:24.991Z', '2023-07-12T18:56:58.797Z', '2022-11-06T04:43:41.065Z', '2023-06-25T20:16:35.829Z', '2022-11-06T04:43:15.230Z', '2023-06-25T10:14:10.501Z', '2022-11-06T04:42:37.244Z', '2023-06-24T20:18:06.564Z', '2022-11-06T04:43:23.649Z', '2022-11-06T04:43:54.939Z', '2023-07-12T15:20:06.187Z', '2022-11-06T13:37:30.065Z', '2023-07-12T16:20:33.418Z', '2022-11-06T04:44:56.893Z', '2023-06-27T18:51:25.229Z', '2022-11-06T14:11:04.269Z', '2023-07-11T19:44:37.745Z', '2022-11-06T14:11:05.403Z', '2022-11-06T04:39:27.016Z', '2023-08-31T08:12:25.314Z', '2022-11-06T04:41:20.365Z', '2023-09-02T23:02:02.944Z', '2022-11-06T04:41:51.551Z', '2022-11-06T04:42:30.449Z', '2023-07-19T01:31:45.362Z', '2022-11-06T13:34:17.157Z', '2022-11-06T04:44:42.815Z', '2023-08-28T20:41:39.160Z', '2022-11-06T04:45:27.038Z', '2022-11-06T04:45:40.737Z', '2022-11-06T04:42:13.735Z', '2022-11-06T04:44:01.906Z', '2022-11-06T04:44:18.606Z', '2023-07-24T18:00:42.669Z', '2022-11-06T04:41:16.496Z', '2022-11-06T04:43:50.358Z', '2023-06-22T15:48:24.193Z', '2022-11-06T04:44:33.791Z', '2023-07-12T16:15:08.739Z', '2022-11-06T04:43:23.012Z', '2023-07-11T21:07:10.196Z', '2022-11-06T04:44:41.906Z', '2023-06-25T12:36:50.125Z', '2022-11-06T04:43:16.594Z', '2023-07-11T20:31:24.516Z', '2022-11-06T04:43:55.701Z', '2023-06-24T11:03:15.691Z', '2022-11-06T14:14:19.319Z', '2023-07-11T18:31:53.385Z', '2022-11-06T13:38:30.103Z', '2022-11-06T13:38:10.854Z', '2022-11-06T04:41:20.609Z', '2023-07-24T18:02:37.722Z', '2022-11-06T04:43:01.273Z', '2022-11-06T04:43:51.058Z', '2022-11-06T04:43:59.275Z', '2022-11-06T13:34:46.222Z', '2023-06-21T13:25:26.939Z', '2022-11-06T04:43:49.287Z', '2022-11-06T04:43:54.186Z', '2022-11-06T04:43:13.303Z', '2023-06-29T19:55:36.667Z', '2023-06-30T07:19:57.427Z', '2022-11-06T04:42:57.429Z', '2022-11-06T04:42:53.943Z'], dtype='<U24') - created(time)<U24'2023-08-30T09:53:24.991Z' ... '...
array(['2023-08-30T09:53:24.991Z', '2023-07-12T18:56:58.797Z', '2022-11-06T04:43:41.065Z', '2023-06-25T20:16:35.829Z', '2022-11-06T04:43:15.230Z', '2023-06-25T10:14:10.501Z', '2022-11-06T04:42:37.244Z', '2023-06-24T20:18:06.564Z', '2022-11-06T04:43:23.649Z', '2022-11-06T04:43:54.939Z', '2023-07-12T15:20:06.187Z', '2022-11-06T13:37:30.065Z', '2023-07-12T16:20:33.418Z', '2022-11-06T04:44:56.893Z', '2023-06-27T18:51:25.229Z', '2022-11-06T14:11:04.269Z', '2023-07-11T19:44:37.745Z', '2022-11-06T14:11:05.403Z', '2022-11-06T04:39:27.016Z', '2023-08-31T08:12:25.314Z', '2022-11-06T04:41:20.365Z', '2023-09-02T23:02:02.944Z', '2022-11-06T04:41:51.551Z', '2022-11-06T04:42:30.449Z', '2023-07-19T01:31:45.362Z', '2022-11-03T10:53:30.936Z', '2022-11-06T04:44:42.815Z', '2023-08-28T20:41:39.160Z', '2022-11-06T04:45:27.038Z', '2022-11-03T10:35:22.039Z', '2022-11-06T04:42:13.735Z', '2022-11-06T04:44:01.906Z', '2022-11-06T04:44:18.606Z', '2023-07-24T18:00:42.669Z', '2022-11-06T04:41:16.496Z', '2022-11-03T10:39:13.068Z', '2023-06-22T15:48:24.193Z', '2022-11-03T10:41:11.108Z', '2023-07-12T16:15:08.739Z', '2022-11-06T04:43:23.012Z', '2023-07-11T21:07:10.196Z', '2022-11-06T04:44:41.906Z', '2023-06-25T12:36:50.125Z', '2022-11-06T04:43:16.594Z', '2023-07-11T20:31:24.516Z', '2022-11-06T04:43:55.701Z', '2023-06-24T11:03:15.691Z', '2022-11-06T14:14:19.319Z', '2023-07-11T18:31:53.385Z', '2022-11-06T13:38:30.103Z', '2022-11-06T13:38:10.854Z', '2022-11-03T10:38:32.333Z', '2023-07-24T18:02:37.722Z', '2022-11-06T04:43:01.273Z', '2022-11-06T04:43:51.058Z', '2022-11-03T10:38:49.526Z', '2022-11-03T10:38:18.195Z', '2023-06-21T13:25:26.939Z', '2022-11-06T04:43:49.287Z', '2022-11-06T04:43:54.186Z', '2022-11-06T04:43:13.303Z', '2023-06-29T19:55:36.667Z', '2023-06-30T07:19:57.427Z', '2022-11-03T10:37:43.273Z', '2022-11-06T04:42:53.943Z'], dtype='<U24') - view:sun_elevation(time)float6420.66 29.41 29.41 ... 19.03 19.33
array([20.65717769, 29.40667789, 29.40655678, 29.53299175, 29.53284968, 31.15131748, 31.15119416, 31.30536438, 31.30522157, 32.98010952, 32.98023196, 34.87277987, 34.87290483, 36.81850237, 36.8186286 , 42.81070593, 43.00682792, 43.00667587, 46.7857159 , 48.86668758, 48.86652367, 54.20732633, 54.20714963, 63.14556905, 62.67508338, 62.67485352, 64.90059226, 64.90080535, 63.79484238, 62.28020247, 61.85137787, 55.9397754 , 51.64576322, 46.66460356, 46.66446865, 44.31518713, 41.29749094, 41.29737536, 40.62479888, 40.62470019, 39.47363892, 39.47353057, 36.916899 , 36.91681058, 35.82282842, 35.82272969, 35.09066166, 34.02427289, 33.29583895, 33.29575963, 32.26912819, 31.55366216, 31.55373753, 29.87006622, 26.7480921 , 25.33890244, 22.16763464, 19.81067577, 19.81062012, 19.81062012, 19.47330287, 19.47335998, 19.03402517, 19.03395836, 19.32933111]) - s2:thin_cirrus_percentage(time)object2.51563 7.028598 ... 0.042559
array([2.51563, 7.028598, 6.062302, 0.00781, 0.120567, 5.96497, 3.32691, 0.024589, 0.037873, 0.018104, 0.000308, 0.070227, 0.069293, 0.004017, 0.018269, 0.754234, 0.002054, 0.000146, 0.371477, 0.428735, 0.464849, 0.027433, 0.041116, 0.221836, 10.13488, 10.168401, 0.022728, 0.463668, 0.426833, 2.168762, 0.519817, 0.002119, 0.426969, 0.387404, 0.06718, 0.027219, 10.991221, 11.106932, 6.085526, 6.020249, 0.030975, 0.009821, 5.873664, 4.157568, 0.003417, 0.001377, 0.994671, 1.056448, 0.000124, 8e-06, 8.33111, 0.000173, 0.010281, 1.1e-05, 0.133605, 5.665855, 0.056168, 0, 0.001398, 0.010097, 1.1e-05, 3.7e-05, 0.000179, 0.000534, 0.042559], dtype=object) - platform(time)<U11'sentinel-2b' ... 'sentinel-2a'
array(['sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2a', 'sentinel-2b', 'sentinel-2a', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a'], dtype='<U11') - s2:water_percentage(time)object0.260838 0.154015 ... 6.159603
array([0.260838, 0.154015, 0.71493, 0.196038, 1.216282, 0.175806, 0.744515, 0.176887, 0.819096, 0.660695, 0.188295, 0.593388, 0.206828, 0.425414, 0.201794, 0.42754, 0.18133, 0.250736, 0.329621, 0.197186, 0.265527, 0.257375, 0.464982, 0.476169, 0.20837, 0.197246, 0.357188, 0.292813, 0.337965, 0.255931, 0.417062, 0.656142, 0.470105, 0.343197, 0.464824, 0.497876, 0.222501, 0.239024, 0.227711, 0.290617, 0.283393, 0.36281, 0.265469, 0.392738, 0.324478, 0.42346, 0.337052, 0.347677, 0.361514, 0.549882, 0.309956, 0.617706, 0.348512, 0.796821, 1.622207, 1.623247, 3.737868, 0.097845, 5.355824, 5.623126, 5.89586, 0.336842, 0.317886, 6.49615, 6.159603], dtype=object) - s2:dark_features_percentage(time)object35.872257 15.490688 ... 33.41836
array([35.872257, 15.490688, 15.426615, 22.757484, 20.31206, 16.900241, 17.298846, 19.806857, 17.954794, 19.344279, 18.605411, 17.298065, 15.841973, 14.723818, 12.47795, 8.147317, 6.486592, 8.368475, 9.136711, 9.471935, 4.877662, 5.215571, 2.829915, 1.553496, 0.970136, 0.609434, 0.754678, 1.151714, 0.941477, 0.45399, 1.702618, 2.958946, 2.909188, 4.941226, 3.938032, 6.376003, 7.052542, 6.861491, 6.710172, 8.241988, 6.533402, 7.349457, 8.667428, 7.683007, 11.972662, 8.467923, 9.474287, 9.115031, 14.991429, 10.522097, 14.116709, 13.383521, 18.600589, 15.634122, 22.90576, 23.055506, 30.60174, 35.566777, 29.157805, 30.762979, 32.97202, 40.977016, 42.140675, 33.06075, 33.41836], dtype=object) - s2:not_vegetated_percentage(time)float646.828 11.64 4.904 ... 6.403 7.516
array([ 6.827541, 11.643875, 4.904069, 12.7378 , 7.811967, 15.13233 , 6.765813, 16.593693, 9.525948, 11.838739, 19.559085, 13.577215, 19.907138, 13.66097 , 22.678789, 12.55991 , 22.722867, 15.128346, 15.931889, 17.465481, 15.302469, 13.346037, 11.371163, 7.183418, 14.248499, 9.690771, 9.223264, 9.58168 , 9.012622, 11.054097, 10.739637, 11.463607, 12.418957, 8.563243, 8.803445, 11.781086, 17.701979, 13.583587, 17.282544, 11.80613 , 13.817489, 10.546423, 8.61038 , 3.60538 , 12.844381, 8.709615, 10.118182, 8.471844, 16.578038, 11.089849, 12.401794, 13.336709, 19.311985, 15.329108, 3.676585, 5.025407, 7.666632, 6.959495, 0.827069, 2.094016, 6.412929, 10.074805, 10.918631, 6.40268 , 7.515902]) - s2:saturated_defective_pixel_percentage()int640
array(0)
- s2:snow_ice_percentage(time)object53.172696 60.569304 ... 23.738836
array([53.172696, 60.569304, 54.313171, 57.653755, 43.74353, 57.544774, 54.037774, 58.4077, 49.270341, 41.84832, 55.127954, 37.863964, 52.323878, 40.777671, 52.27167, 44.621375, 53.832424, 47.302374, 43.722436, 46.102425, 30.143577, 45.875186, 41.257432, 27.877113, 31.303981, 27.278802, 16.500069, 19.468927, 6.136174, 4.367699, 2.55006, 1.795862, 1.572138, 1.543902, 1.469219, 1.82899, 2.796105, 2.337531, 1.900938, 1.758422, 2.897349, 2.63615, 17.610456, 17.242524, 10.428437, 7.091185, 12.082797, 5.790321, 8.214249, 6.919226, 5.038564, 4.362707, 5.516725, 3.702908, 25.313136, 27.462873, 20.145309, 54.500091, 49.163428, 42.097259, 29.398009, 39.025801, 36.363328, 31.210962, 23.738836], dtype=object) - mgrs:grid_square()<U2'PS'
array('PS', dtype='<U2') - s2:cloud_shadow_percentage(time)object0.00411 0.022445 ... 5.073084
array([0.00411, 0.022445, 4.427284, 0.025826, 6.299554, 0.214973, 6.460869, 0.001082, 6.243091, 7.125632, 0.122177, 5.562175, 0.08056, 6.258563, 0.126759, 5.441923, 0.001311, 4.380004, 3.740508, 0.305424, 3.038363, 0.807523, 1.894731, 2.053169, 0.123971, 0.265839, 0.556963, 0.533688, 1.637513, 0.44463, 2.726647, 1.115958, 2.338978, 5.527373, 5.125841, 5.936027, 0.01861, 0.911201, 3.208879, 3.736543, 7.158039, 6.067993, 2.205732, 4.499008, 0.815014, 3.411923, 5.359535, 1.869579, 0.001064, 2.300628, 2.528195, 2.946776, 0.841954, 1.919591, 3.649843, 5.309501, 3.069539, 0.080606, 1.95125, 4.199561, 5.022087, 0.07016, 0, 4.618132, 5.073084], dtype=object) - mgrs:utm_zone()int6432
array(32)
- s2:high_proba_clouds_percentage(time)float640.03288 0.2854 0.479 ... 1.436 1.42
array([3.288100e-02, 2.853590e-01, 4.790390e-01, 6.841700e-02, 3.495720e-01, 1.601230e-01, 3.120390e-01, 1.794600e-02, 3.890670e-01, 3.898030e-01, 2.427900e-02, 5.392310e-01, 1.919700e-02, 1.045976e+00, 5.211030e-01, 8.722986e+00, 6.423000e-03, 4.751880e-01, 4.534660e-01, 5.822150e-01, 1.291065e+00, 2.829218e+00, 3.218969e+00, 9.469271e+00, 1.310789e+00, 1.869626e+00, 4.018103e+00, 2.916400e+00, 9.307959e+00, 6.247902e+00, 9.086780e+00, 1.890319e+00, 5.183531e+00, 7.280029e+00, 7.486678e+00, 9.287885e+00, 3.458050e-01, 5.616310e-01, 3.436800e+00, 3.498174e+00, 7.280371e+00, 7.562798e+00, 3.932654e+00, 5.505205e+00, 6.547320e-01, 1.499930e+00, 7.433846e+00, 3.173025e+00, 3.402000e-03, 5.562020e-01, 6.764350e-01, 7.671570e-01, 3.380080e-01, 3.988670e-01, 4.167500e-01, 1.170431e+00, 1.315025e+00, 3.612000e-03, 1.365308e+00, 1.818938e+00, 1.222941e+00, 1.896200e-02, 1.032000e-03, 1.436024e+00, 1.420145e+00]) - earthsearch:boa_offset_applied(time)boolTrue True False ... False False
array([ True, True, False, True, False, True, False, True, False, False, True, False, True, False, True, False, True, False, False, True, False, True, False, False, True, False, False, True, False, False, False, False, False, True, False, False, True, False, True, False, True, False, True, False, True, False, True, False, True, False, False, False, True, False, False, False, False, True, False, False, False, True, True, False, False]) - s2:medium_proba_clouds_percentage(time)float640.3084 2.001 4.83 ... 0.7785 6.036
array([3.0839200e-01, 2.0012220e+00, 4.8300210e+00, 1.1400400e-01, 7.8954940e+00, 5.4519200e-01, 8.7230500e-01, 7.8470000e-03, 3.4971970e+00, 6.4493430e+00, 3.2733000e-02, 6.7670320e+00, 1.0240000e-02, 6.0864860e+00, 7.0511600e-01, 2.6245750e+00, 1.1135000e-02, 1.1267800e+00, 4.5657100e-01, 6.3280500e-01, 1.2255834e+01, 2.1078810e+00, 2.8801950e+00, 1.7762980e+00, 1.1998870e+00, 2.8858000e+00, 1.4034870e+00, 2.2938500e+00, 2.1778990e+00, 3.4028490e+00, 3.4398040e+00, 1.0365550e+00, 2.0237860e+00, 5.4056610e+00, 1.7999580e+00, 2.4495680e+00, 8.4237900e-01, 1.2673150e+00, 3.3312150e+00, 1.9414730e+00, 5.5801570e+00, 2.3358590e+00, 4.6721980e+00, 3.7431200e+00, 7.7943700e-01, 2.7029620e+00, 5.7600820e+00, 6.6311990e+00, 4.4880000e-03, 6.2181100e-01, 4.2853410e+00, 9.1303400e-01, 3.8397000e-01, 9.6128100e-01, 1.1558286e+01, 3.2200780e+00, 4.9589600e-01, 7.2393000e-02, 2.5707010e+00, 2.1360510e+00, 4.1729000e+00, 1.2638000e-02, 1.4300000e-03, 7.7850800e-01, 6.0363960e+00]) - s2:vegetation_percentage(time)object1.003821 2.674679 ... 11.594474
array([1.003821, 2.674679, 2.938587, 6.434379, 7.419121, 3.186883, 3.323415, 4.962473, 5.664953, 6.822482, 6.334234, 12.347518, 11.537604, 11.502437, 10.984971, 11.278326, 16.755292, 17.688406, 20.000146, 24.586424, 26.839373, 29.225379, 31.2345, 45.217824, 40.248781, 40.689981, 64.543813, 63.144523, 67.6287, 68.127602, 64.335382, 76.903182, 69.595343, 65.299129, 68.121457, 57.554764, 59.904116, 60.774791, 56.98514, 58.596641, 55.844522, 58.502585, 46.137369, 47.877368, 62.104517, 63.009542, 47.673592, 58.927053, 59.844887, 61.083966, 47.538799, 56.53919, 54.613954, 53.221428, 25.320077, 21.666245, 27.581957, 2.711944, 3.611166, 5.17522, 10.205209, 9.482228, 10.255733, 11.158184, 11.594474], dtype=object) - s2:degraded_msi_data_percentage(time)object0.0223 0.025 0 ... 0.0039 0 0
array([0.0223, 0.025, 0, 0.0109, 0, 0.0224, 0, 0.0036, 0, 0, 0.0248, 0, 0.0231, 0, 0.0253, 0, 0.0051, 0, 0, 0.0362, 0, 0.0063, 0, 0, 0.0112, 0, 0, 0.0258, 0, 0, 0, 0, 0, 0.0059, 0, 0, 0.0108, 0, 0.0228, 0, 0.0039, 0, 0.0231, 0, 0.0037, 0, 0.0255, 0, 0.0231, 0, 0, 0, 0.0248, 0, 0, 0, 0, 0.0212, 0, 0, 0, 0.0223, 0.0039, 0, 0], dtype=object) - earthsearch:payload_id(time)<U74'roda-sentinel2/workflow-sentine...
array(['roda-sentinel2/workflow-sentinel2-to-stac/5a93f0464cc0edb8bdbbb2e7a0d2012c', 'roda-sentinel2/workflow-sentinel2-to-stac/563d331b120fee62a7f57075ebf1e004', 'roda-sentinel2/workflow-sentinel2-to-stac/598a8dec1a4123515f6b43db8c007b59', 'roda-sentinel2/workflow-sentinel2-to-stac/ff90e0b59a441d85c80dbe6fe8328957', 'roda-sentinel2/workflow-sentinel2-to-stac/98e1e06f2b2cb73eba246847a0d9b450', 'roda-sentinel2/workflow-sentinel2-to-stac/5f6f0871feb3962cd329be439c10181d', 'roda-sentinel2/workflow-sentinel2-to-stac/b950235c00b037b0f2c2148ecf0b73c7', 'roda-sentinel2/workflow-sentinel2-to-stac/4307d1487cea4f5ea795cdeea4b0fe27', 'roda-sentinel2/workflow-sentinel2-to-stac/0dd09c964b00a39216d78790364a8608', 'roda-sentinel2/workflow-sentinel2-to-stac/7ccce16aef92f0603f08f81d50f614e2', 'roda-sentinel2/workflow-sentinel2-to-stac/b39506708ba27f0f1a2b202a45bc81c0', 'roda-sentinel2/workflow-sentinel2-to-stac/21a009e0f4c71ab77557d121b4cde709', 'roda-sentinel2/workflow-sentinel2-to-stac/1d2fa679bc2794443c175233cf2e782b', 'roda-sentinel2/workflow-sentinel2-to-stac/e76bf62e0487c87d6331bce1274d3761', 'roda-sentinel2/workflow-sentinel2-to-stac/80cead34f3d3c48dcff95581129900db', 'roda-sentinel2/workflow-sentinel2-to-stac/97b157af4bdd0cc477a46d0d4ea5a860', 'roda-sentinel2/workflow-sentinel2-to-stac/811b5b0e61369b4b9b25c806a92e38e0', 'roda-sentinel2/workflow-sentinel2-to-stac/f45fdf5598dede3a3cef5a9cdb461956', 'roda-sentinel2/workflow-sentinel2-to-stac/43fc5027ff60449d117dce677c8e8135', 'roda-sentinel2/workflow-sentinel2-to-stac/4e8597d802361fd111afa86628f46741', ... 'roda-sentinel2/workflow-sentinel2-to-stac/14fde5b1530a5a0f03635435b4f9448d', 'roda-sentinel2/workflow-sentinel2-to-stac/c6f1bedd4fd9719da92f05a68da25171', 'roda-sentinel2/workflow-sentinel2-to-stac/0eb2997c603c34848e43e909afdbbd1a', 'roda-sentinel2/workflow-sentinel2-to-stac/e8f2a7d7b54105b01a394c0e85008e67', 'roda-sentinel2/workflow-sentinel2-to-stac/409bbd2ef9e61f4dec621304c7623221', 'roda-sentinel2/workflow-sentinel2-to-stac/b0bbdcb5a67e9ff1a7aa9b53a80cc8d6', 'roda-sentinel2/workflow-sentinel2-to-stac/ede35147739fd520ee13448b61b3e45c', 'roda-sentinel2/workflow-sentinel2-to-stac/1a3184f69dd0cadcfbd0df48f1392b9f', 'roda-sentinel2/workflow-sentinel2-to-stac/9c05d77aa1eff3eb9f854a50c035550b', 'roda-sentinel2/workflow-sentinel2-to-stac/7ab374294204a80b9bef95d5583df9a6', 'roda-sentinel2/workflow-sentinel2-to-stac/3153efafe0c48eccb67f4657900bf6b9', 'roda-sentinel2/workflow-sentinel2-to-stac/93568ff358f842879ea566c2e766d45d', 'roda-sentinel2/workflow-sentinel2-to-stac/71aba90ac7b3a173c126c68065ea5637', 'roda-sentinel2/workflow-sentinel2-to-stac/8366e35ded0e5bd249b6580f59a52c49', 'roda-sentinel2/workflow-sentinel2-to-stac/ad99eb1e478397b6092c8f33bfb2155a', 'roda-sentinel2/workflow-sentinel2-to-stac/62fdb0869502d7847df53e10633177e6', 'roda-sentinel2/workflow-sentinel2-to-stac/a60d1d53197ebe894647143a010a1bb6', 'roda-sentinel2/workflow-sentinel2-to-stac/313a6f02840be6a2663a0e3230e357b2', 'roda-sentinel2/workflow-sentinel2-to-stac/c821731deb57cc090c7762e61fba8553'], dtype='<U74') - gsd()int6410
array(10)
- proj:shape()object{10980}
array({10980}, dtype=object) - proj:transform()object{0, 600000, 10, 5200020, -10}
array({0, 600000, 10, 5200020, -10}, dtype=object) - title(band)<U20'Red (band 4) - 10m' 'NIR 1 (ban...
array(['Red (band 4) - 10m', 'NIR 1 (band 8) - 10m'], dtype='<U20')
- common_name(band)<U3'red' 'nir'
array(['red', 'nir'], dtype='<U3')
- center_wavelength(band)float640.665 0.842
array([0.665, 0.842])
- full_width_half_max(band)float640.038 0.145
array([0.038, 0.145])
- epsg()int6432632
array(32632)
- timePandasIndex
PandasIndex(DatetimeIndex(['2021-01-11 10:27:54.911000', '2021-02-15 10:27:54.551000', '2021-02-15 10:27:54.552000', '2021-02-17 10:17:57.062000', '2021-02-17 10:17:57.062000', '2021-02-20 10:27:53.334000', '2021-02-20 10:27:53.335000', '2021-02-22 10:17:58.873000', '2021-02-22 10:17:58.873000', '2021-02-25 10:27:55.240000', '2021-02-25 10:27:55.241000', '2021-03-02 10:27:54.438000', '2021-03-02 10:27:54.439000', '2021-03-07 10:27:55.298000', '2021-03-07 10:27:55.300000', '2021-03-22 10:27:54.580000', '2021-03-24 10:17:57.711000', '2021-03-24 10:17:57.712000', '2021-04-01 10:27:53.659000', '2021-04-08 10:17:56.404000', '2021-04-08 10:17:56.404000', '2021-04-23 10:17:55.224000', '2021-04-23 10:17:55.227000', '2021-05-26 10:27:56.205000', '2021-05-28 10:17:59.882000', '2021-05-28 10:17:59.884000', '2021-06-15 10:27:55.829000', '2021-06-15 10:27:55.830000', '2021-06-27 10:18:00.078000', '2021-07-12 10:18:02.331000', '2021-07-20 10:27:57.564000', '2021-08-14 10:27:58.017000', '2021-08-26 10:17:57.394000', '2021-09-10 10:17:59.484000', '2021-09-10 10:17:59.484000', '2021-09-18 10:27:51.968000', '2021-09-25 10:17:57.577000', '2021-09-25 10:17:57.578000', '2021-09-28 10:27:54.449000', '2021-09-28 10:27:54.449000', '2021-09-30 10:18:03.604000', '2021-09-30 10:18:03.604000', '2021-10-08 10:27:56.160000', '2021-10-08 10:27:56.160000', '2021-10-10 10:18:04.559000', '2021-10-10 10:18:04.560000', '2021-10-13 10:28:00.872000', '2021-10-15 10:18:00.588000', '2021-10-18 10:27:56.971000', '2021-10-18 10:27:56.973000', '2021-10-20 10:18:04.648000', '2021-10-23 10:28:00.726000', '2021-10-23 10:28:00.728000', '2021-10-28 10:27:56.929000', '2021-11-07 10:27:55.834000', '2021-11-12 10:27:57.394000', '2021-11-24 10:17:55.331000', '2021-12-12 10:27:49.590000', '2021-12-12 10:27:49.591000', '2021-12-12 10:27:56.297000', '2021-12-17 10:27:49.270000', '2021-12-17 10:27:49.271000', '2021-12-19 10:17:59.960000', '2021-12-19 10:17:59.962000', '2021-12-22 10:27:56.749000'], dtype='datetime64[ns]', name='time', freq=None)) - bandPandasIndex
PandasIndex(Index(['red', 'nir'], dtype='object', name='band'))
We can combine this spatial reducer with the previous over bands to compute a time series of NDVI values:
ndvi_spatial_xr = NDVI(datacube_spatial_median)
ndvi_spatial_xr
<xarray.DataArray 'stackstac-7710f08aba1196bc4f45cd73b40041b2' (time: 65)>
dask.array<truediv, shape=(65,), dtype=float64, chunksize=(1,), chunktype=numpy.ndarray>
Coordinates: (12/46)
* time (time) datetime64[ns] 2021-01-11...
id (time) <U24 'S2B_32TPS_20210111_...
s2:unclassified_percentage (time) object 0.001827 ... 5.000642
s2:product_type <U7 'S2MSI2A'
s2:sequence (time) <U1 '1' '3' '0' ... '0' '0'
s2:generation_time (time) <U27 '2023-05-31T05:43:03...
... ...
s2:degraded_msi_data_percentage (time) object 0.0223 0.025 ... 0 0
earthsearch:payload_id (time) <U74 'roda-sentinel2/work...
gsd int64 10
proj:shape object {10980}
proj:transform object {0, 600000, 10, 5200020, ...
epsg int64 32632- time: 65
- dask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 520 B 8 B Shape (65,) (1,) Dask graph 65 chunks in 12 graph layers Data type float64 numpy.ndarray - time(time)datetime64[ns]2021-01-11T10:27:54.911000 ... 2...
array(['2021-01-11T10:27:54.911000000', '2021-02-15T10:27:54.551000000', '2021-02-15T10:27:54.552000000', '2021-02-17T10:17:57.062000000', '2021-02-17T10:17:57.062000000', '2021-02-20T10:27:53.334000000', '2021-02-20T10:27:53.335000000', '2021-02-22T10:17:58.873000000', '2021-02-22T10:17:58.873000000', '2021-02-25T10:27:55.240000000', '2021-02-25T10:27:55.241000000', '2021-03-02T10:27:54.438000000', '2021-03-02T10:27:54.439000000', '2021-03-07T10:27:55.298000000', '2021-03-07T10:27:55.300000000', '2021-03-22T10:27:54.580000000', '2021-03-24T10:17:57.711000000', '2021-03-24T10:17:57.712000000', '2021-04-01T10:27:53.659000000', '2021-04-08T10:17:56.404000000', '2021-04-08T10:17:56.404000000', '2021-04-23T10:17:55.224000000', '2021-04-23T10:17:55.227000000', '2021-05-26T10:27:56.205000000', '2021-05-28T10:17:59.882000000', '2021-05-28T10:17:59.884000000', '2021-06-15T10:27:55.829000000', '2021-06-15T10:27:55.830000000', '2021-06-27T10:18:00.078000000', '2021-07-12T10:18:02.331000000', '2021-07-20T10:27:57.564000000', '2021-08-14T10:27:58.017000000', '2021-08-26T10:17:57.394000000', '2021-09-10T10:17:59.484000000', '2021-09-10T10:17:59.484000000', '2021-09-18T10:27:51.968000000', '2021-09-25T10:17:57.577000000', '2021-09-25T10:17:57.578000000', '2021-09-28T10:27:54.449000000', '2021-09-28T10:27:54.449000000', '2021-09-30T10:18:03.604000000', '2021-09-30T10:18:03.604000000', '2021-10-08T10:27:56.160000000', '2021-10-08T10:27:56.160000000', '2021-10-10T10:18:04.559000000', '2021-10-10T10:18:04.560000000', '2021-10-13T10:28:00.872000000', '2021-10-15T10:18:00.588000000', '2021-10-18T10:27:56.971000000', '2021-10-18T10:27:56.973000000', '2021-10-20T10:18:04.648000000', '2021-10-23T10:28:00.726000000', '2021-10-23T10:28:00.728000000', '2021-10-28T10:27:56.929000000', '2021-11-07T10:27:55.834000000', '2021-11-12T10:27:57.394000000', '2021-11-24T10:17:55.331000000', '2021-12-12T10:27:49.590000000', '2021-12-12T10:27:49.591000000', '2021-12-12T10:27:56.297000000', '2021-12-17T10:27:49.270000000', '2021-12-17T10:27:49.271000000', '2021-12-19T10:17:59.960000000', '2021-12-19T10:17:59.962000000', '2021-12-22T10:27:56.749000000'], dtype='datetime64[ns]') - id(time)<U24'S2B_32TPS_20210111_1_L2A' ... '...
array(['S2B_32TPS_20210111_1_L2A', 'S2A_32TPS_20210215_3_L2A', 'S2A_32TPS_20210215_0_L2A', 'S2B_32TPS_20210217_1_L2A', 'S2B_32TPS_20210217_0_L2A', 'S2B_32TPS_20210220_1_L2A', 'S2B_32TPS_20210220_0_L2A', 'S2A_32TPS_20210222_1_L2A', 'S2A_32TPS_20210222_0_L2A', 'S2A_32TPS_20210225_0_L2A', 'S2A_32TPS_20210225_3_L2A', 'S2B_32TPS_20210302_0_L2A', 'S2B_32TPS_20210302_1_L2A', 'S2A_32TPS_20210307_0_L2A', 'S2A_32TPS_20210307_1_L2A', 'S2B_32TPS_20210322_0_L2A', 'S2A_32TPS_20210324_1_L2A', 'S2A_32TPS_20210324_0_L2A', 'S2B_32TPS_20210401_0_L2A', 'S2B_32TPS_20210408_2_L2A', 'S2B_32TPS_20210408_0_L2A', 'S2A_32TPS_20210423_2_L2A', 'S2A_32TPS_20210423_0_L2A', 'S2A_32TPS_20210526_0_L2A', 'S2B_32TPS_20210528_1_L2A', 'S2B_32TPS_20210528_0_L2A', 'S2A_32TPS_20210615_0_L2A', 'S2A_32TPS_20210615_1_L2A', 'S2B_32TPS_20210627_0_L2A', 'S2A_32TPS_20210712_0_L2A', 'S2B_32TPS_20210720_0_L2A', 'S2A_32TPS_20210814_0_L2A', 'S2B_32TPS_20210826_0_L2A', 'S2A_32TPS_20210910_1_L2A', 'S2A_32TPS_20210910_0_L2A', 'S2B_32TPS_20210918_0_L2A', 'S2B_32TPS_20210925_1_L2A', 'S2B_32TPS_20210925_0_L2A', 'S2B_32TPS_20210928_1_L2A', 'S2B_32TPS_20210928_0_L2A', 'S2A_32TPS_20210930_1_L2A', 'S2A_32TPS_20210930_0_L2A', 'S2B_32TPS_20211008_1_L2A', 'S2B_32TPS_20211008_0_L2A', 'S2A_32TPS_20211010_1_L2A', 'S2A_32TPS_20211010_0_L2A', 'S2A_32TPS_20211013_2_L2A', 'S2B_32TPS_20211015_0_L2A', 'S2B_32TPS_20211018_1_L2A', 'S2B_32TPS_20211018_0_L2A', 'S2A_32TPS_20211020_0_L2A', 'S2A_32TPS_20211023_0_L2A', 'S2A_32TPS_20211023_3_L2A', 'S2B_32TPS_20211028_0_L2A', 'S2B_32TPS_20211107_0_L2A', 'S2A_32TPS_20211112_1_L2A', 'S2B_32TPS_20211124_0_L2A', 'S2A_32TPS_20211212_2_L2A', 'S2A_32TPS_20211212_1_L2A', 'S2A_32TPS_20211212_0_L2A', 'S2B_32TPS_20211217_0_L2A', 'S2B_32TPS_20211217_1_L2A', 'S2A_32TPS_20211219_1_L2A', 'S2A_32TPS_20211219_0_L2A', 'S2A_32TPS_20211222_0_L2A'], dtype='<U24') - s2:unclassified_percentage(time)object0.001827 0.129823 ... 5.000642
array([0.001827, 0.129823, 5.903982, 0.004486, 4.831852, 0.174702, 6.857514, 0.000936, 6.597646, 5.502601, 0.005526, 5.38119, 0.003286, 5.514648, 0.013578, 5.421813, 0.000571, 5.279551, 5.857176, 0.227375, 5.52128, 0.308397, 4.807, 4.171408, 0.250706, 6.3441, 2.619704, 0.152735, 2.392862, 3.476528, 4.482188, 2.17731, 3.061006, 0.708835, 2.723365, 4.260587, 0.124747, 2.356505, 0.831079, 4.109764, 0.574301, 4.626107, 2.024649, 5.294078, 0.072926, 4.682079, 0.765953, 4.617824, 0.000803, 6.356327, 4.773093, 7.133023, 0.034025, 8.035865, 5.403754, 5.800857, 5.32987, 0.007234, 5.996048, 6.082753, 4.698034, 0.001508, 0.001105, 4.838076, 5.000642], dtype=object) - s2:product_type()<U7'S2MSI2A'
array('S2MSI2A', dtype='<U7') - s2:sequence(time)<U1'1' '3' '0' '1' ... '1' '1' '0' '0'
array(['1', '3', '0', '1', '0', '1', '0', '1', '0', '0', '3', '0', '1', '0', '1', '0', '1', '0', '0', '2', '0', '2', '0', '0', '1', '0', '0', '1', '0', '0', '0', '0', '0', '1', '0', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '2', '0', '1', '0', '0', '0', '3', '0', '0', '1', '0', '2', '1', '0', '0', '1', '1', '0', '0'], dtype='<U1') - s2:generation_time(time)<U27'2023-05-31T05:43:03.000000Z' .....
array(['2023-05-31T05:43:03.000000Z', '2023-06-09T01:27:37.000000Z', '2021-02-15T13:11:02.000000Z', '2023-05-21T16:56:36.000000Z', '2021-02-17T12:09:10.000000Z', '2023-05-19T05:06:36.000000Z', '2021-02-20T13:09:38.000000Z', '2023-05-21T12:51:13.000000Z', '2021-02-22T12:01:41.000000Z', '2021-02-25T13:12:30.000000Z', '2023-06-04T11:19:35.000000Z', '2021-03-02T13:08:05.000000Z', '2023-06-06T02:35:53.000000Z', '2021-03-07T13:16:14.000000Z', '2023-05-25T12:31:49.000000Z', '2021-03-22T13:56:15.000000Z', '2023-06-08T01:43:47.000000Z', '2021-03-24T14:34:56.000000Z', '2021-04-01T14:19:13.000000Z', '2023-05-13T12:44:55.000000Z', '2021-04-08T13:26:17.000000Z', '2023-05-12T19:45:03.000000Z', '2021-04-23T12:55:30.000000Z', '2021-05-26T13:34:25.000000Z', '2023-02-27T08:23:12.000000Z', '2021-05-28T13:08:36.000000Z', '2021-06-15T13:16:59.000000Z', '2023-03-21T22:10:50.000000Z', '2021-06-27T13:58:56.000000Z', '2021-07-12T13:22:19.000000Z', '2021-07-20T13:22:07.000000Z', '2021-08-14T13:23:26.000000Z', '2021-08-26T13:11:06.000000Z', '2023-01-16T19:33:32.000000Z', '2021-09-10T13:19:06.000000Z', '2021-09-18T13:51:51.000000Z', '2023-01-21T20:42:23.000000Z', '2021-09-25T13:47:01.000000Z', '2023-01-25T15:55:19.000000Z', '2021-09-28T13:49:45.000000Z', '2023-01-15T19:24:58.000000Z', '2021-09-30T11:58:00.000000Z', '2023-01-12T12:46:37.000000Z', '2021-10-08T13:24:26.000000Z', '2023-01-13T16:47:44.000000Z', '2021-10-10T13:19:25.000000Z', '2023-01-15T05:24:55.000000Z', '2021-10-15T13:16:08.000000Z', '2023-01-11T00:14:02.000000Z', '2021-10-18T13:18:32.000000Z', '2021-10-20T12:04:20.000000Z', '2021-10-23T13:24:00.000000Z', '2023-01-12T12:45:05.000000Z', '2021-10-28T12:19:42.000000Z', '2021-11-07T12:32:50.000000Z', '2021-11-12T13:19:01.000000Z', '2021-11-24T12:26:23.000000Z', '2022-12-24T05:25:49.000000Z', '2021-12-12T11:50:21.000000Z', '2021-12-12T13:17:17.000000Z', '2021-12-17T12:22:56.000000Z', '2022-12-28T16:01:51.000000Z', '2022-12-27T16:30:48.000000Z', '2021-12-19T12:06:54.000000Z', '2021-12-22T13:16:13.000000Z'], dtype='<U27') - s2:datatake_type()<U8'INS-NOBS'
array('INS-NOBS', dtype='<U8') - proj:epsg()int6432632
array(32632)
- mgrs:latitude_band()<U1'T'
array('T', dtype='<U1') - processing:software(time)object{'sentinel2-to-stac': '0.1.1'} ....
array([{'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}], dtype=object) - s2:processing_baseline(time)<U5'05.00' '05.00' ... '03.01' '03.01'
array(['05.00', '05.00', '02.14', '05.00', '02.14', '05.00', '02.14', '05.00', '02.14', '02.14', '05.00', '02.14', '05.00', '02.14', '05.00', '02.14', '05.00', '02.14', '03.00', '05.00', '03.00', '05.00', '03.00', '03.00', '05.00', '03.00', '03.00', '05.00', '03.00', '03.01', '03.01', '03.01', '03.01', '05.00', '03.01', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '03.01', '03.01', '05.00', '03.01', '03.01', '03.01', '03.01', '05.00', '03.01', '03.01', '03.01', '05.00', '05.00', '03.01', '03.01'], dtype='<U5') - s2:datastrip_id(time)<U64'S2B_OPER_MSI_L2A_DS_S2RP_202305...
array(['S2B_OPER_MSI_L2A_DS_S2RP_20230531T054303_S20210111T102336_N05.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20230609T012737_S20210215T102741_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210215T131102_S20210215T102741_N02.14', 'S2B_OPER_MSI_L2A_DS_S2RP_20230521T165636_S20210217T101023_N05.00', 'S2B_OPER_MSI_L2A_DS_EPAE_20210217T120910_S20210217T101023_N02.14', 'S2B_OPER_MSI_L2A_DS_S2RP_20230519T050636_S20210220T102440_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20210220T130938_S20210220T102440_N02.14', 'S2A_OPER_MSI_L2A_DS_S2RP_20230521T125113_S20210222T101415_N05.00', 'S2A_OPER_MSI_L2A_DS_EPAE_20210222T120141_S20210222T101415_N02.14', 'S2A_OPER_MSI_L2A_DS_VGS2_20210225T131230_S20210225T102740_N02.14', 'S2A_OPER_MSI_L2A_DS_S2RP_20230604T111935_S20210225T102740_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20210302T130805_S20210302T101835_N02.14', 'S2B_OPER_MSI_L2A_DS_S2RP_20230606T023553_S20210302T101835_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210307T131614_S20210307T102135_N02.14', 'S2A_OPER_MSI_L2A_DS_S2RP_20230525T123149_S20210307T102135_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS4_20210322T135615_S20210322T102636_N02.14', 'S2A_OPER_MSI_L2A_DS_S2RP_20230608T014347_S20210324T101606_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210324T143456_S20210324T101606_N02.14', 'S2B_OPER_MSI_L2A_DS_VGS4_20210401T141913_S20210401T102326_N03.00', 'S2B_OPER_MSI_L2A_DS_S2RP_20230513T124455_S20210408T101148_N05.00', ... 'S2A_OPER_MSI_L2A_DS_S2RP_20230115T052455_S20211013T102746_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20211015T131608_S20211015T101504_N03.01', 'S2B_OPER_MSI_L2A_DS_S2RP_20230111T001402_S20211018T102105_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20211018T131832_S20211018T102105_N03.01', 'S2A_OPER_MSI_L2A_DS_VGS4_20211020T120420_S20211020T101421_N03.01', 'S2A_OPER_MSI_L2A_DS_VGS2_20211023T132400_S20211023T102744_N03.01', 'S2A_OPER_MSI_L2A_DS_S2RP_20230112T124505_S20211023T102744_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS4_20211028T121942_S20211028T102038_N03.01', 'S2B_OPER_MSI_L2A_DS_VGS4_20211107T123250_S20211107T102157_N03.01', 'S2A_OPER_MSI_L2A_DS_VGS2_20211112T131901_S20211112T102742_N03.01', 'S2B_OPER_MSI_L2A_DS_VGS4_20211124T122623_S20211124T101240_N03.01', 'S2A_OPER_MSI_L2A_DS_S2RP_20221224T052549_S20211212T102427_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS4_20211212T115021_S20211212T102427_N03.01', 'S2A_OPER_MSI_L2A_DS_VGS2_20211212T131717_S20211212T102745_N03.01', 'S2B_OPER_MSI_L2A_DS_VGS4_20211217T122256_S20211217T102332_N03.01', 'S2B_OPER_MSI_L2A_DS_S2RP_20221228T160151_S20211217T102332_N05.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20221227T163048_S20211219T101431_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS4_20211219T120654_S20211219T101431_N03.01', 'S2A_OPER_MSI_L2A_DS_VGS2_20211222T131613_S20211222T102744_N03.01'], dtype='<U64') - s2:product_uri(time)<U65'S2B_MSIL2A_20210111T102309_N050...
array(['S2B_MSIL2A_20210111T102309_N0500_R065_T32TPS_20230531T054303.SAFE', 'S2A_MSIL2A_20210215T102121_N0500_R065_T32TPS_20230609T012737.SAFE', 'S2A_MSIL2A_20210215T102121_N0214_R065_T32TPS_20210215T131102.SAFE', 'S2B_MSIL2A_20210217T101029_N0500_R022_T32TPS_20230521T165636.SAFE', 'S2B_MSIL2A_20210217T101029_N0214_R022_T32TPS_20210217T120910.SAFE', 'S2B_MSIL2A_20210220T101939_N0500_R065_T32TPS_20230519T050636.SAFE', 'S2B_MSIL2A_20210220T101939_N0214_R065_T32TPS_20210220T130938.SAFE', 'S2A_MSIL2A_20210222T101021_N0500_R022_T32TPS_20230521T125113.SAFE', 'S2A_MSIL2A_20210222T101021_N0214_R022_T32TPS_20210222T120141.SAFE', 'S2A_MSIL2A_20210225T102021_N0214_R065_T32TPS_20210225T131230.SAFE', 'S2A_MSIL2A_20210225T102021_N0500_R065_T32TPS_20230604T111935.SAFE', 'S2B_MSIL2A_20210302T101839_N0214_R065_T32TPS_20210302T130805.SAFE', 'S2B_MSIL2A_20210302T101839_N0500_R065_T32TPS_20230606T023553.SAFE', 'S2A_MSIL2A_20210307T102021_N0214_R065_T32TPS_20210307T131614.SAFE', 'S2A_MSIL2A_20210307T102021_N0500_R065_T32TPS_20230525T123149.SAFE', 'S2B_MSIL2A_20210322T101649_N0214_R065_T32TPS_20210322T135615.SAFE', 'S2A_MSIL2A_20210324T101021_N0500_R022_T32TPS_20230608T014347.SAFE', 'S2A_MSIL2A_20210324T101021_N0214_R022_T32TPS_20210324T143456.SAFE', 'S2B_MSIL2A_20210401T101559_N0300_R065_T32TPS_20210401T141913.SAFE', 'S2B_MSIL2A_20210408T100549_N0500_R022_T32TPS_20230513T124455.SAFE', ... 'S2A_MSIL2A_20211013T101951_N0500_R065_T32TPS_20230115T052455.SAFE', 'S2B_MSIL2A_20211015T101029_N0301_R022_T32TPS_20211015T131608.SAFE', 'S2B_MSIL2A_20211018T101939_N0500_R065_T32TPS_20230111T001402.SAFE', 'S2B_MSIL2A_20211018T101939_N0301_R065_T32TPS_20211018T131832.SAFE', 'S2A_MSIL2A_20211020T101041_N0301_R022_T32TPS_20211020T120420.SAFE', 'S2A_MSIL2A_20211023T102101_N0301_R065_T32TPS_20211023T132400.SAFE', 'S2A_MSIL2A_20211023T102101_N0500_R065_T32TPS_20230112T124505.SAFE', 'S2B_MSIL2A_20211028T102039_N0301_R065_T32TPS_20211028T121942.SAFE', 'S2B_MSIL2A_20211107T102129_N0301_R065_T32TPS_20211107T123250.SAFE', 'S2A_MSIL2A_20211112T102251_N0301_R065_T32TPS_20211112T131901.SAFE', 'S2B_MSIL2A_20211124T101239_N0301_R022_T32TPS_20211124T122623.SAFE', 'S2A_MSIL2A_20211212T102431_N0500_R065_T32TPS_20221224T052549.SAFE', 'S2A_MSIL2A_20211212T102431_N0301_R065_T32TPS_20211212T115021.SAFE', 'S2A_MSIL2A_20211212T102431_N0301_R065_T32TPS_20211212T131717.SAFE', 'S2B_MSIL2A_20211217T102329_N0301_R065_T32TPS_20211217T122256.SAFE', 'S2B_MSIL2A_20211217T102329_N0500_R065_T32TPS_20221228T160151.SAFE', 'S2A_MSIL2A_20211219T101431_N0500_R022_T32TPS_20221227T163048.SAFE', 'S2A_MSIL2A_20211219T101431_N0301_R022_T32TPS_20211219T120654.SAFE', 'S2A_MSIL2A_20211222T102441_N0301_R065_T32TPS_20211222T131613.SAFE'], dtype='<U65') - eo:cloud_cover(time)float642.857 9.315 11.37 ... 2.215 7.499
array([2.8569030e+00, 9.3151780e+00, 1.1371362e+01, 1.9023200e-01, 8.3656320e+00, 6.6702850e+00, 4.5112540e+00, 5.0381000e-02, 3.9241370e+00, 6.8572500e+00, 5.7320000e-02, 7.3764900e+00, 9.8729000e-02, 7.1364790e+00, 1.2444880e+00, 1.2101794e+01, 1.9612000e-02, 1.6021150e+00, 1.2815130e+00, 1.6437540e+00, 1.4011749e+01, 4.9645330e+00, 6.1402800e+00, 1.1467405e+01, 1.2645555e+01, 1.4923827e+01, 5.4443190e+00, 5.6739180e+00, 1.1912692e+01, 1.1819513e+01, 1.3046401e+01, 2.9289930e+00, 7.6342870e+00, 1.3073093e+01, 9.3538160e+00, 1.1764671e+01, 1.2179405e+01, 1.2935878e+01, 1.2853540e+01, 1.1459897e+01, 1.2891503e+01, 9.9084770e+00, 1.4478517e+01, 1.3405893e+01, 1.4375870e+00, 4.2042680e+00, 1.4188600e+01, 1.0860672e+01, 8.0150000e-03, 1.1780210e+00, 1.3292886e+01, 1.6803640e+00, 7.3226000e-01, 1.3601600e+00, 1.2108641e+01, 1.0056364e+01, 1.8670890e+00, 7.6005000e-02, 3.9374070e+00, 3.9650860e+00, 5.3958520e+00, 3.1637000e-02, 2.6410000e-03, 2.2150660e+00, 7.4991000e+00]) - instruments()<U3'msi'
array('msi', dtype='<U3') - s2:datatake_id(time)<U34'GS2B_20210111T102309_020111_N05...
array(['GS2B_20210111T102309_020111_N05.00', 'GS2A_20210215T102121_029520_N05.00', 'GS2A_20210215T102121_029520_N02.14', 'GS2B_20210217T101029_020640_N05.00', 'GS2B_20210217T101029_020640_N02.14', 'GS2B_20210220T101939_020683_N05.00', 'GS2B_20210220T101939_020683_N02.14', 'GS2A_20210222T101021_029620_N05.00', 'GS2A_20210222T101021_029620_N02.14', 'GS2A_20210225T102021_029663_N02.14', 'GS2A_20210225T102021_029663_N05.00', 'GS2B_20210302T101839_020826_N02.14', 'GS2B_20210302T101839_020826_N05.00', 'GS2A_20210307T102021_029806_N02.14', 'GS2A_20210307T102021_029806_N05.00', 'GS2B_20210322T101649_021112_N02.14', 'GS2A_20210324T101021_030049_N05.00', 'GS2A_20210324T101021_030049_N02.14', 'GS2B_20210401T101559_021255_N03.00', 'GS2B_20210408T100549_021355_N05.00', ... 'GS2A_20211010T100941_032909_N03.01', 'GS2A_20211013T101951_032952_N05.00', 'GS2B_20211015T101029_024072_N03.01', 'GS2B_20211018T101939_024115_N05.00', 'GS2B_20211018T101939_024115_N03.01', 'GS2A_20211020T101041_033052_N03.01', 'GS2A_20211023T102101_033095_N03.01', 'GS2A_20211023T102101_033095_N05.00', 'GS2B_20211028T102039_024258_N03.01', 'GS2B_20211107T102129_024401_N03.01', 'GS2A_20211112T102251_033381_N03.01', 'GS2B_20211124T101239_024644_N03.01', 'GS2A_20211212T102431_033810_N05.00', 'GS2A_20211212T102431_033810_N03.01', 'GS2A_20211212T102431_033810_N03.01', 'GS2B_20211217T102329_024973_N03.01', 'GS2B_20211217T102329_024973_N05.00', 'GS2A_20211219T101431_033910_N05.00', 'GS2A_20211219T101431_033910_N03.01', 'GS2A_20211222T102441_033953_N03.01'], dtype='<U34') - s2:nodata_pixel_percentage(time)object11.356236 12.297063 ... 12.545553
array([11.356236, 12.297063, 12.25481, 6.6e-05, 0.000202, 11.384106, 11.153702, 1e-05, 0.000143, 11.5767, 11.612403, 11.321751, 11.543947, 11.545821, 11.591096, 11.302146, 1.3e-05, 0.000182, 11.211243, 2.3e-05, 7.3e-05, 0.005421, 0.002943, 11.126715, 0, 7e-06, 10.792472, 10.827903, 7e-06, 3e-06, 11.535111, 11.207345, 3e-05, 0.00077, 0.000232, 10.601236, 0, 1e-05, 11.48045, 11.250049, 3e-06, 0.000166, 11.772808, 11.547185, 7e-06, 0.000292, 11.806361, 0.000196, 12.032601, 11.805047, 8.3e-05, 11.74587, 11.77223, 12.020504, 11.874768, 11.338881, 0.000448, 66.107917, 66.058993, 27.93659, 10.693993, 10.921072, 4.3e-05, 0.000471, 12.545553], dtype=object) - view:sun_azimuth(time)float64166.1 162.5 162.5 ... 166.6 168.7
array([166.11122138, 162.54877766, 162.54796386, 159.67290702, 159.67208988, 162.22097915, 162.2201451 , 159.31353071, 159.31270178, 161.95804731, 161.95888786, 161.72957356, 161.73043893, 161.54885084, 161.54974115, 161.1584357 , 157.7958327 , 157.79488336, 160.95467327, 157.11384099, 157.11279214, 156.0632289 , 156.06206564, 156.23770514, 151.07526534, 151.07388572, 152.94501975, 152.94641223, 146.80884882, 146.89062175, 152.21378548, 156.8151133 , 155.88046704, 159.86303814, 159.86238306, 165.21262305, 163.40604336, 163.40545498, 167.26091541, 167.26033544, 164.4675583 , 164.46699671, 168.95169412, 168.95114043, 166.23954491, 166.23900125, 169.66922933, 166.94828604, 170.24027851, 170.23974912, 167.58818255, 170.7407418 , 170.74126393, 171.09431652, 171.49581873, 171.5431442 , 168.70337048, 169.79854654, 169.79808633, 169.79808633, 169.2357253 , 169.23619963, 166.6458662 , 166.64539797, 168.6820382 ]) - earthsearch:s3_path(time)<U80's3://sentinel-cogs/sentinel-s2-...
array(['s3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/1/S2B_32TPS_20210111_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2A_32TPS_20210215_3_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2A_32TPS_20210215_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2B_32TPS_20210217_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2B_32TPS_20210217_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2B_32TPS_20210220_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2B_32TPS_20210220_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2A_32TPS_20210222_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2A_32TPS_20210222_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2A_32TPS_20210225_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2A_32TPS_20210225_3_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/3/S2B_32TPS_20210302_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/3/S2B_32TPS_20210302_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/3/S2A_32TPS_20210307_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/3/S2A_32TPS_20210307_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/3/S2B_32TPS_20210322_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/3/S2A_32TPS_20210324_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/3/S2A_32TPS_20210324_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/4/S2B_32TPS_20210401_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/4/S2B_32TPS_20210408_2_L2A', ... 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2A_32TPS_20211013_2_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2B_32TPS_20211015_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2B_32TPS_20211018_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2B_32TPS_20211018_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2A_32TPS_20211020_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2A_32TPS_20211023_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2A_32TPS_20211023_3_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2B_32TPS_20211028_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/11/S2B_32TPS_20211107_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/11/S2A_32TPS_20211112_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/11/S2B_32TPS_20211124_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211212_2_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211212_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211212_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2B_32TPS_20211217_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2B_32TPS_20211217_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211219_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211219_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211222_0_L2A'], dtype='<U80') - constellation()<U10'sentinel-2'
array('sentinel-2', dtype='<U10') - s2:granule_id(time)<U62'S2B_OPER_MSI_L2A_TL_S2RP_202305...
array(['S2B_OPER_MSI_L2A_TL_S2RP_20230531T054303_A020111_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20230609T012737_A029520_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210215T131102_A029520_T32TPS_N02.14', 'S2B_OPER_MSI_L2A_TL_S2RP_20230521T165636_A020640_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_EPAE_20210217T120910_A020640_T32TPS_N02.14', 'S2B_OPER_MSI_L2A_TL_S2RP_20230519T050636_A020683_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20210220T130938_A020683_T32TPS_N02.14', 'S2A_OPER_MSI_L2A_TL_S2RP_20230521T125113_A029620_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_EPAE_20210222T120141_A029620_T32TPS_N02.14', 'S2A_OPER_MSI_L2A_TL_VGS2_20210225T131230_A029663_T32TPS_N02.14', 'S2A_OPER_MSI_L2A_TL_S2RP_20230604T111935_A029663_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20210302T130805_A020826_T32TPS_N02.14', 'S2B_OPER_MSI_L2A_TL_S2RP_20230606T023553_A020826_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210307T131614_A029806_T32TPS_N02.14', 'S2A_OPER_MSI_L2A_TL_S2RP_20230525T123149_A029806_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS4_20210322T135615_A021112_T32TPS_N02.14', 'S2A_OPER_MSI_L2A_TL_S2RP_20230608T014347_A030049_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210324T143456_A030049_T32TPS_N02.14', 'S2B_OPER_MSI_L2A_TL_VGS4_20210401T141913_A021255_T32TPS_N03.00', 'S2B_OPER_MSI_L2A_TL_S2RP_20230513T124455_A021355_T32TPS_N05.00', ... 'S2A_OPER_MSI_L2A_TL_S2RP_20230115T052455_A032952_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20211015T131608_A024072_T32TPS_N03.01', 'S2B_OPER_MSI_L2A_TL_S2RP_20230111T001402_A024115_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20211018T131832_A024115_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_VGS4_20211020T120420_A033052_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_VGS2_20211023T132400_A033095_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_S2RP_20230112T124505_A033095_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS4_20211028T121942_A024258_T32TPS_N03.01', 'S2B_OPER_MSI_L2A_TL_VGS4_20211107T123250_A024401_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_VGS2_20211112T131901_A033381_T32TPS_N03.01', 'S2B_OPER_MSI_L2A_TL_VGS4_20211124T122623_A024644_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_S2RP_20221224T052549_A033810_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS4_20211212T115021_A033810_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_VGS2_20211212T131717_A033810_T32TPS_N03.01', 'S2B_OPER_MSI_L2A_TL_VGS4_20211217T122256_A024973_T32TPS_N03.01', 'S2B_OPER_MSI_L2A_TL_S2RP_20221228T160151_A024973_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20221227T163048_A033910_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS4_20211219T120654_A033910_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_VGS2_20211222T131613_A033953_T32TPS_N03.01'], dtype='<U62') - s2:reflectance_conversion_factor(time)float641.034 1.027 1.027 ... 1.033 1.033
array([1.03421211, 1.02653718, 1.02653718, 1.02577207, 1.02577207, 1.02456125, 1.02456125, 1.0237225 , 1.0237225 , 1.02240605, 1.02240605, 1.02009057, 1.02009057, 1.01763162, 1.01763162, 1.00960406, 1.0084828 , 1.0084828 , 1.00391722, 0.99989016, 0.99989016, 0.99141059, 0.99141059, 0.97577919, 0.97504789, 0.97504789, 0.96985123, 0.96985123, 0.96794362, 0.96743179, 0.96802315, 0.97358286, 0.97804701, 0.98495124, 0.98495124, 0.98910253, 0.9929235 , 0.9929235 , 0.99460618, 0.99460618, 0.99573409, 0.99573409, 1.00031599, 1.00031599, 1.00146456, 1.00146456, 1.00319682, 1.00434323, 1.0060657 , 1.0060657 , 1.00720174, 1.00890228, 1.00890228, 1.01168368, 1.01699809, 1.01948942, 1.02486761, 1.03092533, 1.03092533, 1.03092533, 1.03209956, 1.03209956, 1.03250105, 1.03250105, 1.0330328 ]) - grid:code()<U10'MGRS-32TPS'
array('MGRS-32TPS', dtype='<U10') - updated(time)<U24'2023-08-30T09:53:24.991Z' ... '...
array(['2023-08-30T09:53:24.991Z', '2023-07-12T18:56:58.797Z', '2022-11-06T04:43:41.065Z', '2023-06-25T20:16:35.829Z', '2022-11-06T04:43:15.230Z', '2023-06-25T10:14:10.501Z', '2022-11-06T04:42:37.244Z', '2023-06-24T20:18:06.564Z', '2022-11-06T04:43:23.649Z', '2022-11-06T04:43:54.939Z', '2023-07-12T15:20:06.187Z', '2022-11-06T13:37:30.065Z', '2023-07-12T16:20:33.418Z', '2022-11-06T04:44:56.893Z', '2023-06-27T18:51:25.229Z', '2022-11-06T14:11:04.269Z', '2023-07-11T19:44:37.745Z', '2022-11-06T14:11:05.403Z', '2022-11-06T04:39:27.016Z', '2023-08-31T08:12:25.314Z', '2022-11-06T04:41:20.365Z', '2023-09-02T23:02:02.944Z', '2022-11-06T04:41:51.551Z', '2022-11-06T04:42:30.449Z', '2023-07-19T01:31:45.362Z', '2022-11-06T13:34:17.157Z', '2022-11-06T04:44:42.815Z', '2023-08-28T20:41:39.160Z', '2022-11-06T04:45:27.038Z', '2022-11-06T04:45:40.737Z', '2022-11-06T04:42:13.735Z', '2022-11-06T04:44:01.906Z', '2022-11-06T04:44:18.606Z', '2023-07-24T18:00:42.669Z', '2022-11-06T04:41:16.496Z', '2022-11-06T04:43:50.358Z', '2023-06-22T15:48:24.193Z', '2022-11-06T04:44:33.791Z', '2023-07-12T16:15:08.739Z', '2022-11-06T04:43:23.012Z', '2023-07-11T21:07:10.196Z', '2022-11-06T04:44:41.906Z', '2023-06-25T12:36:50.125Z', '2022-11-06T04:43:16.594Z', '2023-07-11T20:31:24.516Z', '2022-11-06T04:43:55.701Z', '2023-06-24T11:03:15.691Z', '2022-11-06T14:14:19.319Z', '2023-07-11T18:31:53.385Z', '2022-11-06T13:38:30.103Z', '2022-11-06T13:38:10.854Z', '2022-11-06T04:41:20.609Z', '2023-07-24T18:02:37.722Z', '2022-11-06T04:43:01.273Z', '2022-11-06T04:43:51.058Z', '2022-11-06T04:43:59.275Z', '2022-11-06T13:34:46.222Z', '2023-06-21T13:25:26.939Z', '2022-11-06T04:43:49.287Z', '2022-11-06T04:43:54.186Z', '2022-11-06T04:43:13.303Z', '2023-06-29T19:55:36.667Z', '2023-06-30T07:19:57.427Z', '2022-11-06T04:42:57.429Z', '2022-11-06T04:42:53.943Z'], dtype='<U24') - created(time)<U24'2023-08-30T09:53:24.991Z' ... '...
array(['2023-08-30T09:53:24.991Z', '2023-07-12T18:56:58.797Z', '2022-11-06T04:43:41.065Z', '2023-06-25T20:16:35.829Z', '2022-11-06T04:43:15.230Z', '2023-06-25T10:14:10.501Z', '2022-11-06T04:42:37.244Z', '2023-06-24T20:18:06.564Z', '2022-11-06T04:43:23.649Z', '2022-11-06T04:43:54.939Z', '2023-07-12T15:20:06.187Z', '2022-11-06T13:37:30.065Z', '2023-07-12T16:20:33.418Z', '2022-11-06T04:44:56.893Z', '2023-06-27T18:51:25.229Z', '2022-11-06T14:11:04.269Z', '2023-07-11T19:44:37.745Z', '2022-11-06T14:11:05.403Z', '2022-11-06T04:39:27.016Z', '2023-08-31T08:12:25.314Z', '2022-11-06T04:41:20.365Z', '2023-09-02T23:02:02.944Z', '2022-11-06T04:41:51.551Z', '2022-11-06T04:42:30.449Z', '2023-07-19T01:31:45.362Z', '2022-11-03T10:53:30.936Z', '2022-11-06T04:44:42.815Z', '2023-08-28T20:41:39.160Z', '2022-11-06T04:45:27.038Z', '2022-11-03T10:35:22.039Z', '2022-11-06T04:42:13.735Z', '2022-11-06T04:44:01.906Z', '2022-11-06T04:44:18.606Z', '2023-07-24T18:00:42.669Z', '2022-11-06T04:41:16.496Z', '2022-11-03T10:39:13.068Z', '2023-06-22T15:48:24.193Z', '2022-11-03T10:41:11.108Z', '2023-07-12T16:15:08.739Z', '2022-11-06T04:43:23.012Z', '2023-07-11T21:07:10.196Z', '2022-11-06T04:44:41.906Z', '2023-06-25T12:36:50.125Z', '2022-11-06T04:43:16.594Z', '2023-07-11T20:31:24.516Z', '2022-11-06T04:43:55.701Z', '2023-06-24T11:03:15.691Z', '2022-11-06T14:14:19.319Z', '2023-07-11T18:31:53.385Z', '2022-11-06T13:38:30.103Z', '2022-11-06T13:38:10.854Z', '2022-11-03T10:38:32.333Z', '2023-07-24T18:02:37.722Z', '2022-11-06T04:43:01.273Z', '2022-11-06T04:43:51.058Z', '2022-11-03T10:38:49.526Z', '2022-11-03T10:38:18.195Z', '2023-06-21T13:25:26.939Z', '2022-11-06T04:43:49.287Z', '2022-11-06T04:43:54.186Z', '2022-11-06T04:43:13.303Z', '2023-06-29T19:55:36.667Z', '2023-06-30T07:19:57.427Z', '2022-11-03T10:37:43.273Z', '2022-11-06T04:42:53.943Z'], dtype='<U24') - view:sun_elevation(time)float6420.66 29.41 29.41 ... 19.03 19.33
array([20.65717769, 29.40667789, 29.40655678, 29.53299175, 29.53284968, 31.15131748, 31.15119416, 31.30536438, 31.30522157, 32.98010952, 32.98023196, 34.87277987, 34.87290483, 36.81850237, 36.8186286 , 42.81070593, 43.00682792, 43.00667587, 46.7857159 , 48.86668758, 48.86652367, 54.20732633, 54.20714963, 63.14556905, 62.67508338, 62.67485352, 64.90059226, 64.90080535, 63.79484238, 62.28020247, 61.85137787, 55.9397754 , 51.64576322, 46.66460356, 46.66446865, 44.31518713, 41.29749094, 41.29737536, 40.62479888, 40.62470019, 39.47363892, 39.47353057, 36.916899 , 36.91681058, 35.82282842, 35.82272969, 35.09066166, 34.02427289, 33.29583895, 33.29575963, 32.26912819, 31.55366216, 31.55373753, 29.87006622, 26.7480921 , 25.33890244, 22.16763464, 19.81067577, 19.81062012, 19.81062012, 19.47330287, 19.47335998, 19.03402517, 19.03395836, 19.32933111]) - s2:thin_cirrus_percentage(time)object2.51563 7.028598 ... 0.042559
array([2.51563, 7.028598, 6.062302, 0.00781, 0.120567, 5.96497, 3.32691, 0.024589, 0.037873, 0.018104, 0.000308, 0.070227, 0.069293, 0.004017, 0.018269, 0.754234, 0.002054, 0.000146, 0.371477, 0.428735, 0.464849, 0.027433, 0.041116, 0.221836, 10.13488, 10.168401, 0.022728, 0.463668, 0.426833, 2.168762, 0.519817, 0.002119, 0.426969, 0.387404, 0.06718, 0.027219, 10.991221, 11.106932, 6.085526, 6.020249, 0.030975, 0.009821, 5.873664, 4.157568, 0.003417, 0.001377, 0.994671, 1.056448, 0.000124, 8e-06, 8.33111, 0.000173, 0.010281, 1.1e-05, 0.133605, 5.665855, 0.056168, 0, 0.001398, 0.010097, 1.1e-05, 3.7e-05, 0.000179, 0.000534, 0.042559], dtype=object) - platform(time)<U11'sentinel-2b' ... 'sentinel-2a'
array(['sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2a', 'sentinel-2b', 'sentinel-2a', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a'], dtype='<U11') - s2:water_percentage(time)object0.260838 0.154015 ... 6.159603
array([0.260838, 0.154015, 0.71493, 0.196038, 1.216282, 0.175806, 0.744515, 0.176887, 0.819096, 0.660695, 0.188295, 0.593388, 0.206828, 0.425414, 0.201794, 0.42754, 0.18133, 0.250736, 0.329621, 0.197186, 0.265527, 0.257375, 0.464982, 0.476169, 0.20837, 0.197246, 0.357188, 0.292813, 0.337965, 0.255931, 0.417062, 0.656142, 0.470105, 0.343197, 0.464824, 0.497876, 0.222501, 0.239024, 0.227711, 0.290617, 0.283393, 0.36281, 0.265469, 0.392738, 0.324478, 0.42346, 0.337052, 0.347677, 0.361514, 0.549882, 0.309956, 0.617706, 0.348512, 0.796821, 1.622207, 1.623247, 3.737868, 0.097845, 5.355824, 5.623126, 5.89586, 0.336842, 0.317886, 6.49615, 6.159603], dtype=object) - s2:dark_features_percentage(time)object35.872257 15.490688 ... 33.41836
array([35.872257, 15.490688, 15.426615, 22.757484, 20.31206, 16.900241, 17.298846, 19.806857, 17.954794, 19.344279, 18.605411, 17.298065, 15.841973, 14.723818, 12.47795, 8.147317, 6.486592, 8.368475, 9.136711, 9.471935, 4.877662, 5.215571, 2.829915, 1.553496, 0.970136, 0.609434, 0.754678, 1.151714, 0.941477, 0.45399, 1.702618, 2.958946, 2.909188, 4.941226, 3.938032, 6.376003, 7.052542, 6.861491, 6.710172, 8.241988, 6.533402, 7.349457, 8.667428, 7.683007, 11.972662, 8.467923, 9.474287, 9.115031, 14.991429, 10.522097, 14.116709, 13.383521, 18.600589, 15.634122, 22.90576, 23.055506, 30.60174, 35.566777, 29.157805, 30.762979, 32.97202, 40.977016, 42.140675, 33.06075, 33.41836], dtype=object) - s2:not_vegetated_percentage(time)float646.828 11.64 4.904 ... 6.403 7.516
array([ 6.827541, 11.643875, 4.904069, 12.7378 , 7.811967, 15.13233 , 6.765813, 16.593693, 9.525948, 11.838739, 19.559085, 13.577215, 19.907138, 13.66097 , 22.678789, 12.55991 , 22.722867, 15.128346, 15.931889, 17.465481, 15.302469, 13.346037, 11.371163, 7.183418, 14.248499, 9.690771, 9.223264, 9.58168 , 9.012622, 11.054097, 10.739637, 11.463607, 12.418957, 8.563243, 8.803445, 11.781086, 17.701979, 13.583587, 17.282544, 11.80613 , 13.817489, 10.546423, 8.61038 , 3.60538 , 12.844381, 8.709615, 10.118182, 8.471844, 16.578038, 11.089849, 12.401794, 13.336709, 19.311985, 15.329108, 3.676585, 5.025407, 7.666632, 6.959495, 0.827069, 2.094016, 6.412929, 10.074805, 10.918631, 6.40268 , 7.515902]) - s2:saturated_defective_pixel_percentage()int640
array(0)
- s2:snow_ice_percentage(time)object53.172696 60.569304 ... 23.738836
array([53.172696, 60.569304, 54.313171, 57.653755, 43.74353, 57.544774, 54.037774, 58.4077, 49.270341, 41.84832, 55.127954, 37.863964, 52.323878, 40.777671, 52.27167, 44.621375, 53.832424, 47.302374, 43.722436, 46.102425, 30.143577, 45.875186, 41.257432, 27.877113, 31.303981, 27.278802, 16.500069, 19.468927, 6.136174, 4.367699, 2.55006, 1.795862, 1.572138, 1.543902, 1.469219, 1.82899, 2.796105, 2.337531, 1.900938, 1.758422, 2.897349, 2.63615, 17.610456, 17.242524, 10.428437, 7.091185, 12.082797, 5.790321, 8.214249, 6.919226, 5.038564, 4.362707, 5.516725, 3.702908, 25.313136, 27.462873, 20.145309, 54.500091, 49.163428, 42.097259, 29.398009, 39.025801, 36.363328, 31.210962, 23.738836], dtype=object) - mgrs:grid_square()<U2'PS'
array('PS', dtype='<U2') - s2:cloud_shadow_percentage(time)object0.00411 0.022445 ... 5.073084
array([0.00411, 0.022445, 4.427284, 0.025826, 6.299554, 0.214973, 6.460869, 0.001082, 6.243091, 7.125632, 0.122177, 5.562175, 0.08056, 6.258563, 0.126759, 5.441923, 0.001311, 4.380004, 3.740508, 0.305424, 3.038363, 0.807523, 1.894731, 2.053169, 0.123971, 0.265839, 0.556963, 0.533688, 1.637513, 0.44463, 2.726647, 1.115958, 2.338978, 5.527373, 5.125841, 5.936027, 0.01861, 0.911201, 3.208879, 3.736543, 7.158039, 6.067993, 2.205732, 4.499008, 0.815014, 3.411923, 5.359535, 1.869579, 0.001064, 2.300628, 2.528195, 2.946776, 0.841954, 1.919591, 3.649843, 5.309501, 3.069539, 0.080606, 1.95125, 4.199561, 5.022087, 0.07016, 0, 4.618132, 5.073084], dtype=object) - mgrs:utm_zone()int6432
array(32)
- s2:high_proba_clouds_percentage(time)float640.03288 0.2854 0.479 ... 1.436 1.42
array([3.288100e-02, 2.853590e-01, 4.790390e-01, 6.841700e-02, 3.495720e-01, 1.601230e-01, 3.120390e-01, 1.794600e-02, 3.890670e-01, 3.898030e-01, 2.427900e-02, 5.392310e-01, 1.919700e-02, 1.045976e+00, 5.211030e-01, 8.722986e+00, 6.423000e-03, 4.751880e-01, 4.534660e-01, 5.822150e-01, 1.291065e+00, 2.829218e+00, 3.218969e+00, 9.469271e+00, 1.310789e+00, 1.869626e+00, 4.018103e+00, 2.916400e+00, 9.307959e+00, 6.247902e+00, 9.086780e+00, 1.890319e+00, 5.183531e+00, 7.280029e+00, 7.486678e+00, 9.287885e+00, 3.458050e-01, 5.616310e-01, 3.436800e+00, 3.498174e+00, 7.280371e+00, 7.562798e+00, 3.932654e+00, 5.505205e+00, 6.547320e-01, 1.499930e+00, 7.433846e+00, 3.173025e+00, 3.402000e-03, 5.562020e-01, 6.764350e-01, 7.671570e-01, 3.380080e-01, 3.988670e-01, 4.167500e-01, 1.170431e+00, 1.315025e+00, 3.612000e-03, 1.365308e+00, 1.818938e+00, 1.222941e+00, 1.896200e-02, 1.032000e-03, 1.436024e+00, 1.420145e+00]) - earthsearch:boa_offset_applied(time)boolTrue True False ... False False
array([ True, True, False, True, False, True, False, True, False, False, True, False, True, False, True, False, True, False, False, True, False, True, False, False, True, False, False, True, False, False, False, False, False, True, False, False, True, False, True, False, True, False, True, False, True, False, True, False, True, False, False, False, True, False, False, False, False, True, False, False, False, True, True, False, False]) - s2:medium_proba_clouds_percentage(time)float640.3084 2.001 4.83 ... 0.7785 6.036
array([3.0839200e-01, 2.0012220e+00, 4.8300210e+00, 1.1400400e-01, 7.8954940e+00, 5.4519200e-01, 8.7230500e-01, 7.8470000e-03, 3.4971970e+00, 6.4493430e+00, 3.2733000e-02, 6.7670320e+00, 1.0240000e-02, 6.0864860e+00, 7.0511600e-01, 2.6245750e+00, 1.1135000e-02, 1.1267800e+00, 4.5657100e-01, 6.3280500e-01, 1.2255834e+01, 2.1078810e+00, 2.8801950e+00, 1.7762980e+00, 1.1998870e+00, 2.8858000e+00, 1.4034870e+00, 2.2938500e+00, 2.1778990e+00, 3.4028490e+00, 3.4398040e+00, 1.0365550e+00, 2.0237860e+00, 5.4056610e+00, 1.7999580e+00, 2.4495680e+00, 8.4237900e-01, 1.2673150e+00, 3.3312150e+00, 1.9414730e+00, 5.5801570e+00, 2.3358590e+00, 4.6721980e+00, 3.7431200e+00, 7.7943700e-01, 2.7029620e+00, 5.7600820e+00, 6.6311990e+00, 4.4880000e-03, 6.2181100e-01, 4.2853410e+00, 9.1303400e-01, 3.8397000e-01, 9.6128100e-01, 1.1558286e+01, 3.2200780e+00, 4.9589600e-01, 7.2393000e-02, 2.5707010e+00, 2.1360510e+00, 4.1729000e+00, 1.2638000e-02, 1.4300000e-03, 7.7850800e-01, 6.0363960e+00]) - s2:vegetation_percentage(time)object1.003821 2.674679 ... 11.594474
array([1.003821, 2.674679, 2.938587, 6.434379, 7.419121, 3.186883, 3.323415, 4.962473, 5.664953, 6.822482, 6.334234, 12.347518, 11.537604, 11.502437, 10.984971, 11.278326, 16.755292, 17.688406, 20.000146, 24.586424, 26.839373, 29.225379, 31.2345, 45.217824, 40.248781, 40.689981, 64.543813, 63.144523, 67.6287, 68.127602, 64.335382, 76.903182, 69.595343, 65.299129, 68.121457, 57.554764, 59.904116, 60.774791, 56.98514, 58.596641, 55.844522, 58.502585, 46.137369, 47.877368, 62.104517, 63.009542, 47.673592, 58.927053, 59.844887, 61.083966, 47.538799, 56.53919, 54.613954, 53.221428, 25.320077, 21.666245, 27.581957, 2.711944, 3.611166, 5.17522, 10.205209, 9.482228, 10.255733, 11.158184, 11.594474], dtype=object) - s2:degraded_msi_data_percentage(time)object0.0223 0.025 0 ... 0.0039 0 0
array([0.0223, 0.025, 0, 0.0109, 0, 0.0224, 0, 0.0036, 0, 0, 0.0248, 0, 0.0231, 0, 0.0253, 0, 0.0051, 0, 0, 0.0362, 0, 0.0063, 0, 0, 0.0112, 0, 0, 0.0258, 0, 0, 0, 0, 0, 0.0059, 0, 0, 0.0108, 0, 0.0228, 0, 0.0039, 0, 0.0231, 0, 0.0037, 0, 0.0255, 0, 0.0231, 0, 0, 0, 0.0248, 0, 0, 0, 0, 0.0212, 0, 0, 0, 0.0223, 0.0039, 0, 0], dtype=object) - earthsearch:payload_id(time)<U74'roda-sentinel2/workflow-sentine...
array(['roda-sentinel2/workflow-sentinel2-to-stac/5a93f0464cc0edb8bdbbb2e7a0d2012c', 'roda-sentinel2/workflow-sentinel2-to-stac/563d331b120fee62a7f57075ebf1e004', 'roda-sentinel2/workflow-sentinel2-to-stac/598a8dec1a4123515f6b43db8c007b59', 'roda-sentinel2/workflow-sentinel2-to-stac/ff90e0b59a441d85c80dbe6fe8328957', 'roda-sentinel2/workflow-sentinel2-to-stac/98e1e06f2b2cb73eba246847a0d9b450', 'roda-sentinel2/workflow-sentinel2-to-stac/5f6f0871feb3962cd329be439c10181d', 'roda-sentinel2/workflow-sentinel2-to-stac/b950235c00b037b0f2c2148ecf0b73c7', 'roda-sentinel2/workflow-sentinel2-to-stac/4307d1487cea4f5ea795cdeea4b0fe27', 'roda-sentinel2/workflow-sentinel2-to-stac/0dd09c964b00a39216d78790364a8608', 'roda-sentinel2/workflow-sentinel2-to-stac/7ccce16aef92f0603f08f81d50f614e2', 'roda-sentinel2/workflow-sentinel2-to-stac/b39506708ba27f0f1a2b202a45bc81c0', 'roda-sentinel2/workflow-sentinel2-to-stac/21a009e0f4c71ab77557d121b4cde709', 'roda-sentinel2/workflow-sentinel2-to-stac/1d2fa679bc2794443c175233cf2e782b', 'roda-sentinel2/workflow-sentinel2-to-stac/e76bf62e0487c87d6331bce1274d3761', 'roda-sentinel2/workflow-sentinel2-to-stac/80cead34f3d3c48dcff95581129900db', 'roda-sentinel2/workflow-sentinel2-to-stac/97b157af4bdd0cc477a46d0d4ea5a860', 'roda-sentinel2/workflow-sentinel2-to-stac/811b5b0e61369b4b9b25c806a92e38e0', 'roda-sentinel2/workflow-sentinel2-to-stac/f45fdf5598dede3a3cef5a9cdb461956', 'roda-sentinel2/workflow-sentinel2-to-stac/43fc5027ff60449d117dce677c8e8135', 'roda-sentinel2/workflow-sentinel2-to-stac/4e8597d802361fd111afa86628f46741', ... 'roda-sentinel2/workflow-sentinel2-to-stac/14fde5b1530a5a0f03635435b4f9448d', 'roda-sentinel2/workflow-sentinel2-to-stac/c6f1bedd4fd9719da92f05a68da25171', 'roda-sentinel2/workflow-sentinel2-to-stac/0eb2997c603c34848e43e909afdbbd1a', 'roda-sentinel2/workflow-sentinel2-to-stac/e8f2a7d7b54105b01a394c0e85008e67', 'roda-sentinel2/workflow-sentinel2-to-stac/409bbd2ef9e61f4dec621304c7623221', 'roda-sentinel2/workflow-sentinel2-to-stac/b0bbdcb5a67e9ff1a7aa9b53a80cc8d6', 'roda-sentinel2/workflow-sentinel2-to-stac/ede35147739fd520ee13448b61b3e45c', 'roda-sentinel2/workflow-sentinel2-to-stac/1a3184f69dd0cadcfbd0df48f1392b9f', 'roda-sentinel2/workflow-sentinel2-to-stac/9c05d77aa1eff3eb9f854a50c035550b', 'roda-sentinel2/workflow-sentinel2-to-stac/7ab374294204a80b9bef95d5583df9a6', 'roda-sentinel2/workflow-sentinel2-to-stac/3153efafe0c48eccb67f4657900bf6b9', 'roda-sentinel2/workflow-sentinel2-to-stac/93568ff358f842879ea566c2e766d45d', 'roda-sentinel2/workflow-sentinel2-to-stac/71aba90ac7b3a173c126c68065ea5637', 'roda-sentinel2/workflow-sentinel2-to-stac/8366e35ded0e5bd249b6580f59a52c49', 'roda-sentinel2/workflow-sentinel2-to-stac/ad99eb1e478397b6092c8f33bfb2155a', 'roda-sentinel2/workflow-sentinel2-to-stac/62fdb0869502d7847df53e10633177e6', 'roda-sentinel2/workflow-sentinel2-to-stac/a60d1d53197ebe894647143a010a1bb6', 'roda-sentinel2/workflow-sentinel2-to-stac/313a6f02840be6a2663a0e3230e357b2', 'roda-sentinel2/workflow-sentinel2-to-stac/c821731deb57cc090c7762e61fba8553'], dtype='<U74') - gsd()int6410
array(10)
- proj:shape()object{10980}
array({10980}, dtype=object) - proj:transform()object{0, 600000, 10, 5200020, -10}
array({0, 600000, 10, 5200020, -10}, dtype=object) - epsg()int6432632
array(32632)
- timePandasIndex
PandasIndex(DatetimeIndex(['2021-01-11 10:27:54.911000', '2021-02-15 10:27:54.551000', '2021-02-15 10:27:54.552000', '2021-02-17 10:17:57.062000', '2021-02-17 10:17:57.062000', '2021-02-20 10:27:53.334000', '2021-02-20 10:27:53.335000', '2021-02-22 10:17:58.873000', '2021-02-22 10:17:58.873000', '2021-02-25 10:27:55.240000', '2021-02-25 10:27:55.241000', '2021-03-02 10:27:54.438000', '2021-03-02 10:27:54.439000', '2021-03-07 10:27:55.298000', '2021-03-07 10:27:55.300000', '2021-03-22 10:27:54.580000', '2021-03-24 10:17:57.711000', '2021-03-24 10:17:57.712000', '2021-04-01 10:27:53.659000', '2021-04-08 10:17:56.404000', '2021-04-08 10:17:56.404000', '2021-04-23 10:17:55.224000', '2021-04-23 10:17:55.227000', '2021-05-26 10:27:56.205000', '2021-05-28 10:17:59.882000', '2021-05-28 10:17:59.884000', '2021-06-15 10:27:55.829000', '2021-06-15 10:27:55.830000', '2021-06-27 10:18:00.078000', '2021-07-12 10:18:02.331000', '2021-07-20 10:27:57.564000', '2021-08-14 10:27:58.017000', '2021-08-26 10:17:57.394000', '2021-09-10 10:17:59.484000', '2021-09-10 10:17:59.484000', '2021-09-18 10:27:51.968000', '2021-09-25 10:17:57.577000', '2021-09-25 10:17:57.578000', '2021-09-28 10:27:54.449000', '2021-09-28 10:27:54.449000', '2021-09-30 10:18:03.604000', '2021-09-30 10:18:03.604000', '2021-10-08 10:27:56.160000', '2021-10-08 10:27:56.160000', '2021-10-10 10:18:04.559000', '2021-10-10 10:18:04.560000', '2021-10-13 10:28:00.872000', '2021-10-15 10:18:00.588000', '2021-10-18 10:27:56.971000', '2021-10-18 10:27:56.973000', '2021-10-20 10:18:04.648000', '2021-10-23 10:28:00.726000', '2021-10-23 10:28:00.728000', '2021-10-28 10:27:56.929000', '2021-11-07 10:27:55.834000', '2021-11-12 10:27:57.394000', '2021-11-24 10:17:55.331000', '2021-12-12 10:27:49.590000', '2021-12-12 10:27:49.591000', '2021-12-12 10:27:56.297000', '2021-12-17 10:27:49.270000', '2021-12-17 10:27:49.271000', '2021-12-19 10:17:59.960000', '2021-12-19 10:17:59.962000', '2021-12-22 10:27:56.749000'], dtype='datetime64[ns]', name='time', freq=None))
ndvi_spatial_xr.load()
<xarray.DataArray 'stackstac-7710f08aba1196bc4f45cd73b40041b2' (time: 65)>
array([1. , 1. , 0.72450176, 1. , 0.69950249,
1. , 0.55233069, 1. , 0.6202058 , 0.63488844,
1. , 0.69789227, 1. , 0.69595376, 1. ,
0.70536829, 1. , 0.71320346, 0.63970998, 1. ,
0.6643002 , 1. , 0.66222645, 0.77107365, 1. ,
0.70346552, 0.81553398, 1. , 0.80752841, 0.83191411,
0.81575657, 0.82283105, 0.84372402, 1. , 0.83625219,
0.81262327, 1. , 0.8295082 , 1. , 0.84407484,
1. , 0.8255814 , 1. , 0.84149419, 1. ,
0.82274742, 1. , 0.70364437, 1. , 0.82560706,
0.77223851, 0.80659341, 1. , 0.80912863, 0.76804734,
0.7275 , 0.77817963, 1. , 0.99874135, 0.86932447,
0.82356729, 1. , 1. , 0.79028436, 0.78220286])
Coordinates: (12/46)
* time (time) datetime64[ns] 2021-01-11...
id (time) <U24 'S2B_32TPS_20210111_...
s2:unclassified_percentage (time) object 0.001827 ... 5.000642
s2:product_type <U7 'S2MSI2A'
s2:sequence (time) <U1 '1' '3' '0' ... '0' '0'
s2:generation_time (time) <U27 '2023-05-31T05:43:03...
... ...
s2:degraded_msi_data_percentage (time) object 0.0223 0.025 ... 0 0
earthsearch:payload_id (time) <U74 'roda-sentinel2/work...
gsd int64 10
proj:shape object {10980}
proj:transform object {0, 600000, 10, 5200020, ...
epsg int64 32632- time: 65
- 1.0 1.0 0.7245 1.0 0.6995 1.0 ... 0.8693 0.8236 1.0 1.0 0.7903 0.7822
array([1. , 1. , 0.72450176, 1. , 0.69950249, 1. , 0.55233069, 1. , 0.6202058 , 0.63488844, 1. , 0.69789227, 1. , 0.69595376, 1. , 0.70536829, 1. , 0.71320346, 0.63970998, 1. , 0.6643002 , 1. , 0.66222645, 0.77107365, 1. , 0.70346552, 0.81553398, 1. , 0.80752841, 0.83191411, 0.81575657, 0.82283105, 0.84372402, 1. , 0.83625219, 0.81262327, 1. , 0.8295082 , 1. , 0.84407484, 1. , 0.8255814 , 1. , 0.84149419, 1. , 0.82274742, 1. , 0.70364437, 1. , 0.82560706, 0.77223851, 0.80659341, 1. , 0.80912863, 0.76804734, 0.7275 , 0.77817963, 1. , 0.99874135, 0.86932447, 0.82356729, 1. , 1. , 0.79028436, 0.78220286]) - time(time)datetime64[ns]2021-01-11T10:27:54.911000 ... 2...
array(['2021-01-11T10:27:54.911000000', '2021-02-15T10:27:54.551000000', '2021-02-15T10:27:54.552000000', '2021-02-17T10:17:57.062000000', '2021-02-17T10:17:57.062000000', '2021-02-20T10:27:53.334000000', '2021-02-20T10:27:53.335000000', '2021-02-22T10:17:58.873000000', '2021-02-22T10:17:58.873000000', '2021-02-25T10:27:55.240000000', '2021-02-25T10:27:55.241000000', '2021-03-02T10:27:54.438000000', '2021-03-02T10:27:54.439000000', '2021-03-07T10:27:55.298000000', '2021-03-07T10:27:55.300000000', '2021-03-22T10:27:54.580000000', '2021-03-24T10:17:57.711000000', '2021-03-24T10:17:57.712000000', '2021-04-01T10:27:53.659000000', '2021-04-08T10:17:56.404000000', '2021-04-08T10:17:56.404000000', '2021-04-23T10:17:55.224000000', '2021-04-23T10:17:55.227000000', '2021-05-26T10:27:56.205000000', '2021-05-28T10:17:59.882000000', '2021-05-28T10:17:59.884000000', '2021-06-15T10:27:55.829000000', '2021-06-15T10:27:55.830000000', '2021-06-27T10:18:00.078000000', '2021-07-12T10:18:02.331000000', '2021-07-20T10:27:57.564000000', '2021-08-14T10:27:58.017000000', '2021-08-26T10:17:57.394000000', '2021-09-10T10:17:59.484000000', '2021-09-10T10:17:59.484000000', '2021-09-18T10:27:51.968000000', '2021-09-25T10:17:57.577000000', '2021-09-25T10:17:57.578000000', '2021-09-28T10:27:54.449000000', '2021-09-28T10:27:54.449000000', '2021-09-30T10:18:03.604000000', '2021-09-30T10:18:03.604000000', '2021-10-08T10:27:56.160000000', '2021-10-08T10:27:56.160000000', '2021-10-10T10:18:04.559000000', '2021-10-10T10:18:04.560000000', '2021-10-13T10:28:00.872000000', '2021-10-15T10:18:00.588000000', '2021-10-18T10:27:56.971000000', '2021-10-18T10:27:56.973000000', '2021-10-20T10:18:04.648000000', '2021-10-23T10:28:00.726000000', '2021-10-23T10:28:00.728000000', '2021-10-28T10:27:56.929000000', '2021-11-07T10:27:55.834000000', '2021-11-12T10:27:57.394000000', '2021-11-24T10:17:55.331000000', '2021-12-12T10:27:49.590000000', '2021-12-12T10:27:49.591000000', '2021-12-12T10:27:56.297000000', '2021-12-17T10:27:49.270000000', '2021-12-17T10:27:49.271000000', '2021-12-19T10:17:59.960000000', '2021-12-19T10:17:59.962000000', '2021-12-22T10:27:56.749000000'], dtype='datetime64[ns]') - id(time)<U24'S2B_32TPS_20210111_1_L2A' ... '...
array(['S2B_32TPS_20210111_1_L2A', 'S2A_32TPS_20210215_3_L2A', 'S2A_32TPS_20210215_0_L2A', 'S2B_32TPS_20210217_1_L2A', 'S2B_32TPS_20210217_0_L2A', 'S2B_32TPS_20210220_1_L2A', 'S2B_32TPS_20210220_0_L2A', 'S2A_32TPS_20210222_1_L2A', 'S2A_32TPS_20210222_0_L2A', 'S2A_32TPS_20210225_0_L2A', 'S2A_32TPS_20210225_3_L2A', 'S2B_32TPS_20210302_0_L2A', 'S2B_32TPS_20210302_1_L2A', 'S2A_32TPS_20210307_0_L2A', 'S2A_32TPS_20210307_1_L2A', 'S2B_32TPS_20210322_0_L2A', 'S2A_32TPS_20210324_1_L2A', 'S2A_32TPS_20210324_0_L2A', 'S2B_32TPS_20210401_0_L2A', 'S2B_32TPS_20210408_2_L2A', 'S2B_32TPS_20210408_0_L2A', 'S2A_32TPS_20210423_2_L2A', 'S2A_32TPS_20210423_0_L2A', 'S2A_32TPS_20210526_0_L2A', 'S2B_32TPS_20210528_1_L2A', 'S2B_32TPS_20210528_0_L2A', 'S2A_32TPS_20210615_0_L2A', 'S2A_32TPS_20210615_1_L2A', 'S2B_32TPS_20210627_0_L2A', 'S2A_32TPS_20210712_0_L2A', 'S2B_32TPS_20210720_0_L2A', 'S2A_32TPS_20210814_0_L2A', 'S2B_32TPS_20210826_0_L2A', 'S2A_32TPS_20210910_1_L2A', 'S2A_32TPS_20210910_0_L2A', 'S2B_32TPS_20210918_0_L2A', 'S2B_32TPS_20210925_1_L2A', 'S2B_32TPS_20210925_0_L2A', 'S2B_32TPS_20210928_1_L2A', 'S2B_32TPS_20210928_0_L2A', 'S2A_32TPS_20210930_1_L2A', 'S2A_32TPS_20210930_0_L2A', 'S2B_32TPS_20211008_1_L2A', 'S2B_32TPS_20211008_0_L2A', 'S2A_32TPS_20211010_1_L2A', 'S2A_32TPS_20211010_0_L2A', 'S2A_32TPS_20211013_2_L2A', 'S2B_32TPS_20211015_0_L2A', 'S2B_32TPS_20211018_1_L2A', 'S2B_32TPS_20211018_0_L2A', 'S2A_32TPS_20211020_0_L2A', 'S2A_32TPS_20211023_0_L2A', 'S2A_32TPS_20211023_3_L2A', 'S2B_32TPS_20211028_0_L2A', 'S2B_32TPS_20211107_0_L2A', 'S2A_32TPS_20211112_1_L2A', 'S2B_32TPS_20211124_0_L2A', 'S2A_32TPS_20211212_2_L2A', 'S2A_32TPS_20211212_1_L2A', 'S2A_32TPS_20211212_0_L2A', 'S2B_32TPS_20211217_0_L2A', 'S2B_32TPS_20211217_1_L2A', 'S2A_32TPS_20211219_1_L2A', 'S2A_32TPS_20211219_0_L2A', 'S2A_32TPS_20211222_0_L2A'], dtype='<U24') - s2:unclassified_percentage(time)object0.001827 0.129823 ... 5.000642
array([0.001827, 0.129823, 5.903982, 0.004486, 4.831852, 0.174702, 6.857514, 0.000936, 6.597646, 5.502601, 0.005526, 5.38119, 0.003286, 5.514648, 0.013578, 5.421813, 0.000571, 5.279551, 5.857176, 0.227375, 5.52128, 0.308397, 4.807, 4.171408, 0.250706, 6.3441, 2.619704, 0.152735, 2.392862, 3.476528, 4.482188, 2.17731, 3.061006, 0.708835, 2.723365, 4.260587, 0.124747, 2.356505, 0.831079, 4.109764, 0.574301, 4.626107, 2.024649, 5.294078, 0.072926, 4.682079, 0.765953, 4.617824, 0.000803, 6.356327, 4.773093, 7.133023, 0.034025, 8.035865, 5.403754, 5.800857, 5.32987, 0.007234, 5.996048, 6.082753, 4.698034, 0.001508, 0.001105, 4.838076, 5.000642], dtype=object) - s2:product_type()<U7'S2MSI2A'
array('S2MSI2A', dtype='<U7') - s2:sequence(time)<U1'1' '3' '0' '1' ... '1' '1' '0' '0'
array(['1', '3', '0', '1', '0', '1', '0', '1', '0', '0', '3', '0', '1', '0', '1', '0', '1', '0', '0', '2', '0', '2', '0', '0', '1', '0', '0', '1', '0', '0', '0', '0', '0', '1', '0', '0', '1', '0', '1', '0', '1', '0', '1', '0', '1', '0', '2', '0', '1', '0', '0', '0', '3', '0', '0', '1', '0', '2', '1', '0', '0', '1', '1', '0', '0'], dtype='<U1') - s2:generation_time(time)<U27'2023-05-31T05:43:03.000000Z' .....
array(['2023-05-31T05:43:03.000000Z', '2023-06-09T01:27:37.000000Z', '2021-02-15T13:11:02.000000Z', '2023-05-21T16:56:36.000000Z', '2021-02-17T12:09:10.000000Z', '2023-05-19T05:06:36.000000Z', '2021-02-20T13:09:38.000000Z', '2023-05-21T12:51:13.000000Z', '2021-02-22T12:01:41.000000Z', '2021-02-25T13:12:30.000000Z', '2023-06-04T11:19:35.000000Z', '2021-03-02T13:08:05.000000Z', '2023-06-06T02:35:53.000000Z', '2021-03-07T13:16:14.000000Z', '2023-05-25T12:31:49.000000Z', '2021-03-22T13:56:15.000000Z', '2023-06-08T01:43:47.000000Z', '2021-03-24T14:34:56.000000Z', '2021-04-01T14:19:13.000000Z', '2023-05-13T12:44:55.000000Z', '2021-04-08T13:26:17.000000Z', '2023-05-12T19:45:03.000000Z', '2021-04-23T12:55:30.000000Z', '2021-05-26T13:34:25.000000Z', '2023-02-27T08:23:12.000000Z', '2021-05-28T13:08:36.000000Z', '2021-06-15T13:16:59.000000Z', '2023-03-21T22:10:50.000000Z', '2021-06-27T13:58:56.000000Z', '2021-07-12T13:22:19.000000Z', '2021-07-20T13:22:07.000000Z', '2021-08-14T13:23:26.000000Z', '2021-08-26T13:11:06.000000Z', '2023-01-16T19:33:32.000000Z', '2021-09-10T13:19:06.000000Z', '2021-09-18T13:51:51.000000Z', '2023-01-21T20:42:23.000000Z', '2021-09-25T13:47:01.000000Z', '2023-01-25T15:55:19.000000Z', '2021-09-28T13:49:45.000000Z', '2023-01-15T19:24:58.000000Z', '2021-09-30T11:58:00.000000Z', '2023-01-12T12:46:37.000000Z', '2021-10-08T13:24:26.000000Z', '2023-01-13T16:47:44.000000Z', '2021-10-10T13:19:25.000000Z', '2023-01-15T05:24:55.000000Z', '2021-10-15T13:16:08.000000Z', '2023-01-11T00:14:02.000000Z', '2021-10-18T13:18:32.000000Z', '2021-10-20T12:04:20.000000Z', '2021-10-23T13:24:00.000000Z', '2023-01-12T12:45:05.000000Z', '2021-10-28T12:19:42.000000Z', '2021-11-07T12:32:50.000000Z', '2021-11-12T13:19:01.000000Z', '2021-11-24T12:26:23.000000Z', '2022-12-24T05:25:49.000000Z', '2021-12-12T11:50:21.000000Z', '2021-12-12T13:17:17.000000Z', '2021-12-17T12:22:56.000000Z', '2022-12-28T16:01:51.000000Z', '2022-12-27T16:30:48.000000Z', '2021-12-19T12:06:54.000000Z', '2021-12-22T13:16:13.000000Z'], dtype='<U27') - s2:datatake_type()<U8'INS-NOBS'
array('INS-NOBS', dtype='<U8') - proj:epsg()int6432632
array(32632)
- mgrs:latitude_band()<U1'T'
array('T', dtype='<U1') - processing:software(time)object{'sentinel2-to-stac': '0.1.1'} ....
array([{'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.1'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}, {'sentinel2-to-stac': '0.1.0'}], dtype=object) - s2:processing_baseline(time)<U5'05.00' '05.00' ... '03.01' '03.01'
array(['05.00', '05.00', '02.14', '05.00', '02.14', '05.00', '02.14', '05.00', '02.14', '02.14', '05.00', '02.14', '05.00', '02.14', '05.00', '02.14', '05.00', '02.14', '03.00', '05.00', '03.00', '05.00', '03.00', '03.00', '05.00', '03.00', '03.00', '05.00', '03.00', '03.01', '03.01', '03.01', '03.01', '05.00', '03.01', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '05.00', '03.01', '03.01', '03.01', '05.00', '03.01', '03.01', '03.01', '03.01', '05.00', '03.01', '03.01', '03.01', '05.00', '05.00', '03.01', '03.01'], dtype='<U5') - s2:datastrip_id(time)<U64'S2B_OPER_MSI_L2A_DS_S2RP_202305...
array(['S2B_OPER_MSI_L2A_DS_S2RP_20230531T054303_S20210111T102336_N05.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20230609T012737_S20210215T102741_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210215T131102_S20210215T102741_N02.14', 'S2B_OPER_MSI_L2A_DS_S2RP_20230521T165636_S20210217T101023_N05.00', 'S2B_OPER_MSI_L2A_DS_EPAE_20210217T120910_S20210217T101023_N02.14', 'S2B_OPER_MSI_L2A_DS_S2RP_20230519T050636_S20210220T102440_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20210220T130938_S20210220T102440_N02.14', 'S2A_OPER_MSI_L2A_DS_S2RP_20230521T125113_S20210222T101415_N05.00', 'S2A_OPER_MSI_L2A_DS_EPAE_20210222T120141_S20210222T101415_N02.14', 'S2A_OPER_MSI_L2A_DS_VGS2_20210225T131230_S20210225T102740_N02.14', 'S2A_OPER_MSI_L2A_DS_S2RP_20230604T111935_S20210225T102740_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20210302T130805_S20210302T101835_N02.14', 'S2B_OPER_MSI_L2A_DS_S2RP_20230606T023553_S20210302T101835_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210307T131614_S20210307T102135_N02.14', 'S2A_OPER_MSI_L2A_DS_S2RP_20230525T123149_S20210307T102135_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS4_20210322T135615_S20210322T102636_N02.14', 'S2A_OPER_MSI_L2A_DS_S2RP_20230608T014347_S20210324T101606_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS2_20210324T143456_S20210324T101606_N02.14', 'S2B_OPER_MSI_L2A_DS_VGS4_20210401T141913_S20210401T102326_N03.00', 'S2B_OPER_MSI_L2A_DS_S2RP_20230513T124455_S20210408T101148_N05.00', ... 'S2A_OPER_MSI_L2A_DS_S2RP_20230115T052455_S20211013T102746_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20211015T131608_S20211015T101504_N03.01', 'S2B_OPER_MSI_L2A_DS_S2RP_20230111T001402_S20211018T102105_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS2_20211018T131832_S20211018T102105_N03.01', 'S2A_OPER_MSI_L2A_DS_VGS4_20211020T120420_S20211020T101421_N03.01', 'S2A_OPER_MSI_L2A_DS_VGS2_20211023T132400_S20211023T102744_N03.01', 'S2A_OPER_MSI_L2A_DS_S2RP_20230112T124505_S20211023T102744_N05.00', 'S2B_OPER_MSI_L2A_DS_VGS4_20211028T121942_S20211028T102038_N03.01', 'S2B_OPER_MSI_L2A_DS_VGS4_20211107T123250_S20211107T102157_N03.01', 'S2A_OPER_MSI_L2A_DS_VGS2_20211112T131901_S20211112T102742_N03.01', 'S2B_OPER_MSI_L2A_DS_VGS4_20211124T122623_S20211124T101240_N03.01', 'S2A_OPER_MSI_L2A_DS_S2RP_20221224T052549_S20211212T102427_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS4_20211212T115021_S20211212T102427_N03.01', 'S2A_OPER_MSI_L2A_DS_VGS2_20211212T131717_S20211212T102745_N03.01', 'S2B_OPER_MSI_L2A_DS_VGS4_20211217T122256_S20211217T102332_N03.01', 'S2B_OPER_MSI_L2A_DS_S2RP_20221228T160151_S20211217T102332_N05.00', 'S2A_OPER_MSI_L2A_DS_S2RP_20221227T163048_S20211219T101431_N05.00', 'S2A_OPER_MSI_L2A_DS_VGS4_20211219T120654_S20211219T101431_N03.01', 'S2A_OPER_MSI_L2A_DS_VGS2_20211222T131613_S20211222T102744_N03.01'], dtype='<U64') - s2:product_uri(time)<U65'S2B_MSIL2A_20210111T102309_N050...
array(['S2B_MSIL2A_20210111T102309_N0500_R065_T32TPS_20230531T054303.SAFE', 'S2A_MSIL2A_20210215T102121_N0500_R065_T32TPS_20230609T012737.SAFE', 'S2A_MSIL2A_20210215T102121_N0214_R065_T32TPS_20210215T131102.SAFE', 'S2B_MSIL2A_20210217T101029_N0500_R022_T32TPS_20230521T165636.SAFE', 'S2B_MSIL2A_20210217T101029_N0214_R022_T32TPS_20210217T120910.SAFE', 'S2B_MSIL2A_20210220T101939_N0500_R065_T32TPS_20230519T050636.SAFE', 'S2B_MSIL2A_20210220T101939_N0214_R065_T32TPS_20210220T130938.SAFE', 'S2A_MSIL2A_20210222T101021_N0500_R022_T32TPS_20230521T125113.SAFE', 'S2A_MSIL2A_20210222T101021_N0214_R022_T32TPS_20210222T120141.SAFE', 'S2A_MSIL2A_20210225T102021_N0214_R065_T32TPS_20210225T131230.SAFE', 'S2A_MSIL2A_20210225T102021_N0500_R065_T32TPS_20230604T111935.SAFE', 'S2B_MSIL2A_20210302T101839_N0214_R065_T32TPS_20210302T130805.SAFE', 'S2B_MSIL2A_20210302T101839_N0500_R065_T32TPS_20230606T023553.SAFE', 'S2A_MSIL2A_20210307T102021_N0214_R065_T32TPS_20210307T131614.SAFE', 'S2A_MSIL2A_20210307T102021_N0500_R065_T32TPS_20230525T123149.SAFE', 'S2B_MSIL2A_20210322T101649_N0214_R065_T32TPS_20210322T135615.SAFE', 'S2A_MSIL2A_20210324T101021_N0500_R022_T32TPS_20230608T014347.SAFE', 'S2A_MSIL2A_20210324T101021_N0214_R022_T32TPS_20210324T143456.SAFE', 'S2B_MSIL2A_20210401T101559_N0300_R065_T32TPS_20210401T141913.SAFE', 'S2B_MSIL2A_20210408T100549_N0500_R022_T32TPS_20230513T124455.SAFE', ... 'S2A_MSIL2A_20211013T101951_N0500_R065_T32TPS_20230115T052455.SAFE', 'S2B_MSIL2A_20211015T101029_N0301_R022_T32TPS_20211015T131608.SAFE', 'S2B_MSIL2A_20211018T101939_N0500_R065_T32TPS_20230111T001402.SAFE', 'S2B_MSIL2A_20211018T101939_N0301_R065_T32TPS_20211018T131832.SAFE', 'S2A_MSIL2A_20211020T101041_N0301_R022_T32TPS_20211020T120420.SAFE', 'S2A_MSIL2A_20211023T102101_N0301_R065_T32TPS_20211023T132400.SAFE', 'S2A_MSIL2A_20211023T102101_N0500_R065_T32TPS_20230112T124505.SAFE', 'S2B_MSIL2A_20211028T102039_N0301_R065_T32TPS_20211028T121942.SAFE', 'S2B_MSIL2A_20211107T102129_N0301_R065_T32TPS_20211107T123250.SAFE', 'S2A_MSIL2A_20211112T102251_N0301_R065_T32TPS_20211112T131901.SAFE', 'S2B_MSIL2A_20211124T101239_N0301_R022_T32TPS_20211124T122623.SAFE', 'S2A_MSIL2A_20211212T102431_N0500_R065_T32TPS_20221224T052549.SAFE', 'S2A_MSIL2A_20211212T102431_N0301_R065_T32TPS_20211212T115021.SAFE', 'S2A_MSIL2A_20211212T102431_N0301_R065_T32TPS_20211212T131717.SAFE', 'S2B_MSIL2A_20211217T102329_N0301_R065_T32TPS_20211217T122256.SAFE', 'S2B_MSIL2A_20211217T102329_N0500_R065_T32TPS_20221228T160151.SAFE', 'S2A_MSIL2A_20211219T101431_N0500_R022_T32TPS_20221227T163048.SAFE', 'S2A_MSIL2A_20211219T101431_N0301_R022_T32TPS_20211219T120654.SAFE', 'S2A_MSIL2A_20211222T102441_N0301_R065_T32TPS_20211222T131613.SAFE'], dtype='<U65') - eo:cloud_cover(time)float642.857 9.315 11.37 ... 2.215 7.499
array([2.8569030e+00, 9.3151780e+00, 1.1371362e+01, 1.9023200e-01, 8.3656320e+00, 6.6702850e+00, 4.5112540e+00, 5.0381000e-02, 3.9241370e+00, 6.8572500e+00, 5.7320000e-02, 7.3764900e+00, 9.8729000e-02, 7.1364790e+00, 1.2444880e+00, 1.2101794e+01, 1.9612000e-02, 1.6021150e+00, 1.2815130e+00, 1.6437540e+00, 1.4011749e+01, 4.9645330e+00, 6.1402800e+00, 1.1467405e+01, 1.2645555e+01, 1.4923827e+01, 5.4443190e+00, 5.6739180e+00, 1.1912692e+01, 1.1819513e+01, 1.3046401e+01, 2.9289930e+00, 7.6342870e+00, 1.3073093e+01, 9.3538160e+00, 1.1764671e+01, 1.2179405e+01, 1.2935878e+01, 1.2853540e+01, 1.1459897e+01, 1.2891503e+01, 9.9084770e+00, 1.4478517e+01, 1.3405893e+01, 1.4375870e+00, 4.2042680e+00, 1.4188600e+01, 1.0860672e+01, 8.0150000e-03, 1.1780210e+00, 1.3292886e+01, 1.6803640e+00, 7.3226000e-01, 1.3601600e+00, 1.2108641e+01, 1.0056364e+01, 1.8670890e+00, 7.6005000e-02, 3.9374070e+00, 3.9650860e+00, 5.3958520e+00, 3.1637000e-02, 2.6410000e-03, 2.2150660e+00, 7.4991000e+00]) - instruments()<U3'msi'
array('msi', dtype='<U3') - s2:datatake_id(time)<U34'GS2B_20210111T102309_020111_N05...
array(['GS2B_20210111T102309_020111_N05.00', 'GS2A_20210215T102121_029520_N05.00', 'GS2A_20210215T102121_029520_N02.14', 'GS2B_20210217T101029_020640_N05.00', 'GS2B_20210217T101029_020640_N02.14', 'GS2B_20210220T101939_020683_N05.00', 'GS2B_20210220T101939_020683_N02.14', 'GS2A_20210222T101021_029620_N05.00', 'GS2A_20210222T101021_029620_N02.14', 'GS2A_20210225T102021_029663_N02.14', 'GS2A_20210225T102021_029663_N05.00', 'GS2B_20210302T101839_020826_N02.14', 'GS2B_20210302T101839_020826_N05.00', 'GS2A_20210307T102021_029806_N02.14', 'GS2A_20210307T102021_029806_N05.00', 'GS2B_20210322T101649_021112_N02.14', 'GS2A_20210324T101021_030049_N05.00', 'GS2A_20210324T101021_030049_N02.14', 'GS2B_20210401T101559_021255_N03.00', 'GS2B_20210408T100549_021355_N05.00', ... 'GS2A_20211010T100941_032909_N03.01', 'GS2A_20211013T101951_032952_N05.00', 'GS2B_20211015T101029_024072_N03.01', 'GS2B_20211018T101939_024115_N05.00', 'GS2B_20211018T101939_024115_N03.01', 'GS2A_20211020T101041_033052_N03.01', 'GS2A_20211023T102101_033095_N03.01', 'GS2A_20211023T102101_033095_N05.00', 'GS2B_20211028T102039_024258_N03.01', 'GS2B_20211107T102129_024401_N03.01', 'GS2A_20211112T102251_033381_N03.01', 'GS2B_20211124T101239_024644_N03.01', 'GS2A_20211212T102431_033810_N05.00', 'GS2A_20211212T102431_033810_N03.01', 'GS2A_20211212T102431_033810_N03.01', 'GS2B_20211217T102329_024973_N03.01', 'GS2B_20211217T102329_024973_N05.00', 'GS2A_20211219T101431_033910_N05.00', 'GS2A_20211219T101431_033910_N03.01', 'GS2A_20211222T102441_033953_N03.01'], dtype='<U34') - s2:nodata_pixel_percentage(time)object11.356236 12.297063 ... 12.545553
array([11.356236, 12.297063, 12.25481, 6.6e-05, 0.000202, 11.384106, 11.153702, 1e-05, 0.000143, 11.5767, 11.612403, 11.321751, 11.543947, 11.545821, 11.591096, 11.302146, 1.3e-05, 0.000182, 11.211243, 2.3e-05, 7.3e-05, 0.005421, 0.002943, 11.126715, 0, 7e-06, 10.792472, 10.827903, 7e-06, 3e-06, 11.535111, 11.207345, 3e-05, 0.00077, 0.000232, 10.601236, 0, 1e-05, 11.48045, 11.250049, 3e-06, 0.000166, 11.772808, 11.547185, 7e-06, 0.000292, 11.806361, 0.000196, 12.032601, 11.805047, 8.3e-05, 11.74587, 11.77223, 12.020504, 11.874768, 11.338881, 0.000448, 66.107917, 66.058993, 27.93659, 10.693993, 10.921072, 4.3e-05, 0.000471, 12.545553], dtype=object) - view:sun_azimuth(time)float64166.1 162.5 162.5 ... 166.6 168.7
array([166.11122138, 162.54877766, 162.54796386, 159.67290702, 159.67208988, 162.22097915, 162.2201451 , 159.31353071, 159.31270178, 161.95804731, 161.95888786, 161.72957356, 161.73043893, 161.54885084, 161.54974115, 161.1584357 , 157.7958327 , 157.79488336, 160.95467327, 157.11384099, 157.11279214, 156.0632289 , 156.06206564, 156.23770514, 151.07526534, 151.07388572, 152.94501975, 152.94641223, 146.80884882, 146.89062175, 152.21378548, 156.8151133 , 155.88046704, 159.86303814, 159.86238306, 165.21262305, 163.40604336, 163.40545498, 167.26091541, 167.26033544, 164.4675583 , 164.46699671, 168.95169412, 168.95114043, 166.23954491, 166.23900125, 169.66922933, 166.94828604, 170.24027851, 170.23974912, 167.58818255, 170.7407418 , 170.74126393, 171.09431652, 171.49581873, 171.5431442 , 168.70337048, 169.79854654, 169.79808633, 169.79808633, 169.2357253 , 169.23619963, 166.6458662 , 166.64539797, 168.6820382 ]) - earthsearch:s3_path(time)<U80's3://sentinel-cogs/sentinel-s2-...
array(['s3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/1/S2B_32TPS_20210111_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2A_32TPS_20210215_3_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2A_32TPS_20210215_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2B_32TPS_20210217_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2B_32TPS_20210217_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2B_32TPS_20210220_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2B_32TPS_20210220_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2A_32TPS_20210222_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2A_32TPS_20210222_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2A_32TPS_20210225_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/2/S2A_32TPS_20210225_3_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/3/S2B_32TPS_20210302_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/3/S2B_32TPS_20210302_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/3/S2A_32TPS_20210307_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/3/S2A_32TPS_20210307_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/3/S2B_32TPS_20210322_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/3/S2A_32TPS_20210324_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/3/S2A_32TPS_20210324_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/4/S2B_32TPS_20210401_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/4/S2B_32TPS_20210408_2_L2A', ... 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2A_32TPS_20211013_2_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2B_32TPS_20211015_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2B_32TPS_20211018_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2B_32TPS_20211018_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2A_32TPS_20211020_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2A_32TPS_20211023_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2A_32TPS_20211023_3_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/10/S2B_32TPS_20211028_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/11/S2B_32TPS_20211107_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/11/S2A_32TPS_20211112_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/11/S2B_32TPS_20211124_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211212_2_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211212_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211212_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2B_32TPS_20211217_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2B_32TPS_20211217_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211219_1_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211219_0_L2A', 's3://sentinel-cogs/sentinel-s2-l2a-cogs/32/T/PS/2021/12/S2A_32TPS_20211222_0_L2A'], dtype='<U80') - constellation()<U10'sentinel-2'
array('sentinel-2', dtype='<U10') - s2:granule_id(time)<U62'S2B_OPER_MSI_L2A_TL_S2RP_202305...
array(['S2B_OPER_MSI_L2A_TL_S2RP_20230531T054303_A020111_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20230609T012737_A029520_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210215T131102_A029520_T32TPS_N02.14', 'S2B_OPER_MSI_L2A_TL_S2RP_20230521T165636_A020640_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_EPAE_20210217T120910_A020640_T32TPS_N02.14', 'S2B_OPER_MSI_L2A_TL_S2RP_20230519T050636_A020683_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20210220T130938_A020683_T32TPS_N02.14', 'S2A_OPER_MSI_L2A_TL_S2RP_20230521T125113_A029620_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_EPAE_20210222T120141_A029620_T32TPS_N02.14', 'S2A_OPER_MSI_L2A_TL_VGS2_20210225T131230_A029663_T32TPS_N02.14', 'S2A_OPER_MSI_L2A_TL_S2RP_20230604T111935_A029663_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20210302T130805_A020826_T32TPS_N02.14', 'S2B_OPER_MSI_L2A_TL_S2RP_20230606T023553_A020826_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210307T131614_A029806_T32TPS_N02.14', 'S2A_OPER_MSI_L2A_TL_S2RP_20230525T123149_A029806_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS4_20210322T135615_A021112_T32TPS_N02.14', 'S2A_OPER_MSI_L2A_TL_S2RP_20230608T014347_A030049_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS2_20210324T143456_A030049_T32TPS_N02.14', 'S2B_OPER_MSI_L2A_TL_VGS4_20210401T141913_A021255_T32TPS_N03.00', 'S2B_OPER_MSI_L2A_TL_S2RP_20230513T124455_A021355_T32TPS_N05.00', ... 'S2A_OPER_MSI_L2A_TL_S2RP_20230115T052455_A032952_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20211015T131608_A024072_T32TPS_N03.01', 'S2B_OPER_MSI_L2A_TL_S2RP_20230111T001402_A024115_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS2_20211018T131832_A024115_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_VGS4_20211020T120420_A033052_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_VGS2_20211023T132400_A033095_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_S2RP_20230112T124505_A033095_T32TPS_N05.00', 'S2B_OPER_MSI_L2A_TL_VGS4_20211028T121942_A024258_T32TPS_N03.01', 'S2B_OPER_MSI_L2A_TL_VGS4_20211107T123250_A024401_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_VGS2_20211112T131901_A033381_T32TPS_N03.01', 'S2B_OPER_MSI_L2A_TL_VGS4_20211124T122623_A024644_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_S2RP_20221224T052549_A033810_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS4_20211212T115021_A033810_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_VGS2_20211212T131717_A033810_T32TPS_N03.01', 'S2B_OPER_MSI_L2A_TL_VGS4_20211217T122256_A024973_T32TPS_N03.01', 'S2B_OPER_MSI_L2A_TL_S2RP_20221228T160151_A024973_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_S2RP_20221227T163048_A033910_T32TPS_N05.00', 'S2A_OPER_MSI_L2A_TL_VGS4_20211219T120654_A033910_T32TPS_N03.01', 'S2A_OPER_MSI_L2A_TL_VGS2_20211222T131613_A033953_T32TPS_N03.01'], dtype='<U62') - s2:reflectance_conversion_factor(time)float641.034 1.027 1.027 ... 1.033 1.033
array([1.03421211, 1.02653718, 1.02653718, 1.02577207, 1.02577207, 1.02456125, 1.02456125, 1.0237225 , 1.0237225 , 1.02240605, 1.02240605, 1.02009057, 1.02009057, 1.01763162, 1.01763162, 1.00960406, 1.0084828 , 1.0084828 , 1.00391722, 0.99989016, 0.99989016, 0.99141059, 0.99141059, 0.97577919, 0.97504789, 0.97504789, 0.96985123, 0.96985123, 0.96794362, 0.96743179, 0.96802315, 0.97358286, 0.97804701, 0.98495124, 0.98495124, 0.98910253, 0.9929235 , 0.9929235 , 0.99460618, 0.99460618, 0.99573409, 0.99573409, 1.00031599, 1.00031599, 1.00146456, 1.00146456, 1.00319682, 1.00434323, 1.0060657 , 1.0060657 , 1.00720174, 1.00890228, 1.00890228, 1.01168368, 1.01699809, 1.01948942, 1.02486761, 1.03092533, 1.03092533, 1.03092533, 1.03209956, 1.03209956, 1.03250105, 1.03250105, 1.0330328 ]) - grid:code()<U10'MGRS-32TPS'
array('MGRS-32TPS', dtype='<U10') - updated(time)<U24'2023-08-30T09:53:24.991Z' ... '...
array(['2023-08-30T09:53:24.991Z', '2023-07-12T18:56:58.797Z', '2022-11-06T04:43:41.065Z', '2023-06-25T20:16:35.829Z', '2022-11-06T04:43:15.230Z', '2023-06-25T10:14:10.501Z', '2022-11-06T04:42:37.244Z', '2023-06-24T20:18:06.564Z', '2022-11-06T04:43:23.649Z', '2022-11-06T04:43:54.939Z', '2023-07-12T15:20:06.187Z', '2022-11-06T13:37:30.065Z', '2023-07-12T16:20:33.418Z', '2022-11-06T04:44:56.893Z', '2023-06-27T18:51:25.229Z', '2022-11-06T14:11:04.269Z', '2023-07-11T19:44:37.745Z', '2022-11-06T14:11:05.403Z', '2022-11-06T04:39:27.016Z', '2023-08-31T08:12:25.314Z', '2022-11-06T04:41:20.365Z', '2023-09-02T23:02:02.944Z', '2022-11-06T04:41:51.551Z', '2022-11-06T04:42:30.449Z', '2023-07-19T01:31:45.362Z', '2022-11-06T13:34:17.157Z', '2022-11-06T04:44:42.815Z', '2023-08-28T20:41:39.160Z', '2022-11-06T04:45:27.038Z', '2022-11-06T04:45:40.737Z', '2022-11-06T04:42:13.735Z', '2022-11-06T04:44:01.906Z', '2022-11-06T04:44:18.606Z', '2023-07-24T18:00:42.669Z', '2022-11-06T04:41:16.496Z', '2022-11-06T04:43:50.358Z', '2023-06-22T15:48:24.193Z', '2022-11-06T04:44:33.791Z', '2023-07-12T16:15:08.739Z', '2022-11-06T04:43:23.012Z', '2023-07-11T21:07:10.196Z', '2022-11-06T04:44:41.906Z', '2023-06-25T12:36:50.125Z', '2022-11-06T04:43:16.594Z', '2023-07-11T20:31:24.516Z', '2022-11-06T04:43:55.701Z', '2023-06-24T11:03:15.691Z', '2022-11-06T14:14:19.319Z', '2023-07-11T18:31:53.385Z', '2022-11-06T13:38:30.103Z', '2022-11-06T13:38:10.854Z', '2022-11-06T04:41:20.609Z', '2023-07-24T18:02:37.722Z', '2022-11-06T04:43:01.273Z', '2022-11-06T04:43:51.058Z', '2022-11-06T04:43:59.275Z', '2022-11-06T13:34:46.222Z', '2023-06-21T13:25:26.939Z', '2022-11-06T04:43:49.287Z', '2022-11-06T04:43:54.186Z', '2022-11-06T04:43:13.303Z', '2023-06-29T19:55:36.667Z', '2023-06-30T07:19:57.427Z', '2022-11-06T04:42:57.429Z', '2022-11-06T04:42:53.943Z'], dtype='<U24') - created(time)<U24'2023-08-30T09:53:24.991Z' ... '...
array(['2023-08-30T09:53:24.991Z', '2023-07-12T18:56:58.797Z', '2022-11-06T04:43:41.065Z', '2023-06-25T20:16:35.829Z', '2022-11-06T04:43:15.230Z', '2023-06-25T10:14:10.501Z', '2022-11-06T04:42:37.244Z', '2023-06-24T20:18:06.564Z', '2022-11-06T04:43:23.649Z', '2022-11-06T04:43:54.939Z', '2023-07-12T15:20:06.187Z', '2022-11-06T13:37:30.065Z', '2023-07-12T16:20:33.418Z', '2022-11-06T04:44:56.893Z', '2023-06-27T18:51:25.229Z', '2022-11-06T14:11:04.269Z', '2023-07-11T19:44:37.745Z', '2022-11-06T14:11:05.403Z', '2022-11-06T04:39:27.016Z', '2023-08-31T08:12:25.314Z', '2022-11-06T04:41:20.365Z', '2023-09-02T23:02:02.944Z', '2022-11-06T04:41:51.551Z', '2022-11-06T04:42:30.449Z', '2023-07-19T01:31:45.362Z', '2022-11-03T10:53:30.936Z', '2022-11-06T04:44:42.815Z', '2023-08-28T20:41:39.160Z', '2022-11-06T04:45:27.038Z', '2022-11-03T10:35:22.039Z', '2022-11-06T04:42:13.735Z', '2022-11-06T04:44:01.906Z', '2022-11-06T04:44:18.606Z', '2023-07-24T18:00:42.669Z', '2022-11-06T04:41:16.496Z', '2022-11-03T10:39:13.068Z', '2023-06-22T15:48:24.193Z', '2022-11-03T10:41:11.108Z', '2023-07-12T16:15:08.739Z', '2022-11-06T04:43:23.012Z', '2023-07-11T21:07:10.196Z', '2022-11-06T04:44:41.906Z', '2023-06-25T12:36:50.125Z', '2022-11-06T04:43:16.594Z', '2023-07-11T20:31:24.516Z', '2022-11-06T04:43:55.701Z', '2023-06-24T11:03:15.691Z', '2022-11-06T14:14:19.319Z', '2023-07-11T18:31:53.385Z', '2022-11-06T13:38:30.103Z', '2022-11-06T13:38:10.854Z', '2022-11-03T10:38:32.333Z', '2023-07-24T18:02:37.722Z', '2022-11-06T04:43:01.273Z', '2022-11-06T04:43:51.058Z', '2022-11-03T10:38:49.526Z', '2022-11-03T10:38:18.195Z', '2023-06-21T13:25:26.939Z', '2022-11-06T04:43:49.287Z', '2022-11-06T04:43:54.186Z', '2022-11-06T04:43:13.303Z', '2023-06-29T19:55:36.667Z', '2023-06-30T07:19:57.427Z', '2022-11-03T10:37:43.273Z', '2022-11-06T04:42:53.943Z'], dtype='<U24') - view:sun_elevation(time)float6420.66 29.41 29.41 ... 19.03 19.33
array([20.65717769, 29.40667789, 29.40655678, 29.53299175, 29.53284968, 31.15131748, 31.15119416, 31.30536438, 31.30522157, 32.98010952, 32.98023196, 34.87277987, 34.87290483, 36.81850237, 36.8186286 , 42.81070593, 43.00682792, 43.00667587, 46.7857159 , 48.86668758, 48.86652367, 54.20732633, 54.20714963, 63.14556905, 62.67508338, 62.67485352, 64.90059226, 64.90080535, 63.79484238, 62.28020247, 61.85137787, 55.9397754 , 51.64576322, 46.66460356, 46.66446865, 44.31518713, 41.29749094, 41.29737536, 40.62479888, 40.62470019, 39.47363892, 39.47353057, 36.916899 , 36.91681058, 35.82282842, 35.82272969, 35.09066166, 34.02427289, 33.29583895, 33.29575963, 32.26912819, 31.55366216, 31.55373753, 29.87006622, 26.7480921 , 25.33890244, 22.16763464, 19.81067577, 19.81062012, 19.81062012, 19.47330287, 19.47335998, 19.03402517, 19.03395836, 19.32933111]) - s2:thin_cirrus_percentage(time)object2.51563 7.028598 ... 0.042559
array([2.51563, 7.028598, 6.062302, 0.00781, 0.120567, 5.96497, 3.32691, 0.024589, 0.037873, 0.018104, 0.000308, 0.070227, 0.069293, 0.004017, 0.018269, 0.754234, 0.002054, 0.000146, 0.371477, 0.428735, 0.464849, 0.027433, 0.041116, 0.221836, 10.13488, 10.168401, 0.022728, 0.463668, 0.426833, 2.168762, 0.519817, 0.002119, 0.426969, 0.387404, 0.06718, 0.027219, 10.991221, 11.106932, 6.085526, 6.020249, 0.030975, 0.009821, 5.873664, 4.157568, 0.003417, 0.001377, 0.994671, 1.056448, 0.000124, 8e-06, 8.33111, 0.000173, 0.010281, 1.1e-05, 0.133605, 5.665855, 0.056168, 0, 0.001398, 0.010097, 1.1e-05, 3.7e-05, 0.000179, 0.000534, 0.042559], dtype=object) - platform(time)<U11'sentinel-2b' ... 'sentinel-2a'
array(['sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2a', 'sentinel-2b', 'sentinel-2a', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a', 'sentinel-2b', 'sentinel-2b', 'sentinel-2a', 'sentinel-2a', 'sentinel-2a'], dtype='<U11') - s2:water_percentage(time)object0.260838 0.154015 ... 6.159603
array([0.260838, 0.154015, 0.71493, 0.196038, 1.216282, 0.175806, 0.744515, 0.176887, 0.819096, 0.660695, 0.188295, 0.593388, 0.206828, 0.425414, 0.201794, 0.42754, 0.18133, 0.250736, 0.329621, 0.197186, 0.265527, 0.257375, 0.464982, 0.476169, 0.20837, 0.197246, 0.357188, 0.292813, 0.337965, 0.255931, 0.417062, 0.656142, 0.470105, 0.343197, 0.464824, 0.497876, 0.222501, 0.239024, 0.227711, 0.290617, 0.283393, 0.36281, 0.265469, 0.392738, 0.324478, 0.42346, 0.337052, 0.347677, 0.361514, 0.549882, 0.309956, 0.617706, 0.348512, 0.796821, 1.622207, 1.623247, 3.737868, 0.097845, 5.355824, 5.623126, 5.89586, 0.336842, 0.317886, 6.49615, 6.159603], dtype=object) - s2:dark_features_percentage(time)object35.872257 15.490688 ... 33.41836
array([35.872257, 15.490688, 15.426615, 22.757484, 20.31206, 16.900241, 17.298846, 19.806857, 17.954794, 19.344279, 18.605411, 17.298065, 15.841973, 14.723818, 12.47795, 8.147317, 6.486592, 8.368475, 9.136711, 9.471935, 4.877662, 5.215571, 2.829915, 1.553496, 0.970136, 0.609434, 0.754678, 1.151714, 0.941477, 0.45399, 1.702618, 2.958946, 2.909188, 4.941226, 3.938032, 6.376003, 7.052542, 6.861491, 6.710172, 8.241988, 6.533402, 7.349457, 8.667428, 7.683007, 11.972662, 8.467923, 9.474287, 9.115031, 14.991429, 10.522097, 14.116709, 13.383521, 18.600589, 15.634122, 22.90576, 23.055506, 30.60174, 35.566777, 29.157805, 30.762979, 32.97202, 40.977016, 42.140675, 33.06075, 33.41836], dtype=object) - s2:not_vegetated_percentage(time)float646.828 11.64 4.904 ... 6.403 7.516
array([ 6.827541, 11.643875, 4.904069, 12.7378 , 7.811967, 15.13233 , 6.765813, 16.593693, 9.525948, 11.838739, 19.559085, 13.577215, 19.907138, 13.66097 , 22.678789, 12.55991 , 22.722867, 15.128346, 15.931889, 17.465481, 15.302469, 13.346037, 11.371163, 7.183418, 14.248499, 9.690771, 9.223264, 9.58168 , 9.012622, 11.054097, 10.739637, 11.463607, 12.418957, 8.563243, 8.803445, 11.781086, 17.701979, 13.583587, 17.282544, 11.80613 , 13.817489, 10.546423, 8.61038 , 3.60538 , 12.844381, 8.709615, 10.118182, 8.471844, 16.578038, 11.089849, 12.401794, 13.336709, 19.311985, 15.329108, 3.676585, 5.025407, 7.666632, 6.959495, 0.827069, 2.094016, 6.412929, 10.074805, 10.918631, 6.40268 , 7.515902]) - s2:saturated_defective_pixel_percentage()int640
array(0)
- s2:snow_ice_percentage(time)object53.172696 60.569304 ... 23.738836
array([53.172696, 60.569304, 54.313171, 57.653755, 43.74353, 57.544774, 54.037774, 58.4077, 49.270341, 41.84832, 55.127954, 37.863964, 52.323878, 40.777671, 52.27167, 44.621375, 53.832424, 47.302374, 43.722436, 46.102425, 30.143577, 45.875186, 41.257432, 27.877113, 31.303981, 27.278802, 16.500069, 19.468927, 6.136174, 4.367699, 2.55006, 1.795862, 1.572138, 1.543902, 1.469219, 1.82899, 2.796105, 2.337531, 1.900938, 1.758422, 2.897349, 2.63615, 17.610456, 17.242524, 10.428437, 7.091185, 12.082797, 5.790321, 8.214249, 6.919226, 5.038564, 4.362707, 5.516725, 3.702908, 25.313136, 27.462873, 20.145309, 54.500091, 49.163428, 42.097259, 29.398009, 39.025801, 36.363328, 31.210962, 23.738836], dtype=object) - mgrs:grid_square()<U2'PS'
array('PS', dtype='<U2') - s2:cloud_shadow_percentage(time)object0.00411 0.022445 ... 5.073084
array([0.00411, 0.022445, 4.427284, 0.025826, 6.299554, 0.214973, 6.460869, 0.001082, 6.243091, 7.125632, 0.122177, 5.562175, 0.08056, 6.258563, 0.126759, 5.441923, 0.001311, 4.380004, 3.740508, 0.305424, 3.038363, 0.807523, 1.894731, 2.053169, 0.123971, 0.265839, 0.556963, 0.533688, 1.637513, 0.44463, 2.726647, 1.115958, 2.338978, 5.527373, 5.125841, 5.936027, 0.01861, 0.911201, 3.208879, 3.736543, 7.158039, 6.067993, 2.205732, 4.499008, 0.815014, 3.411923, 5.359535, 1.869579, 0.001064, 2.300628, 2.528195, 2.946776, 0.841954, 1.919591, 3.649843, 5.309501, 3.069539, 0.080606, 1.95125, 4.199561, 5.022087, 0.07016, 0, 4.618132, 5.073084], dtype=object) - mgrs:utm_zone()int6432
array(32)
- s2:high_proba_clouds_percentage(time)float640.03288 0.2854 0.479 ... 1.436 1.42
array([3.288100e-02, 2.853590e-01, 4.790390e-01, 6.841700e-02, 3.495720e-01, 1.601230e-01, 3.120390e-01, 1.794600e-02, 3.890670e-01, 3.898030e-01, 2.427900e-02, 5.392310e-01, 1.919700e-02, 1.045976e+00, 5.211030e-01, 8.722986e+00, 6.423000e-03, 4.751880e-01, 4.534660e-01, 5.822150e-01, 1.291065e+00, 2.829218e+00, 3.218969e+00, 9.469271e+00, 1.310789e+00, 1.869626e+00, 4.018103e+00, 2.916400e+00, 9.307959e+00, 6.247902e+00, 9.086780e+00, 1.890319e+00, 5.183531e+00, 7.280029e+00, 7.486678e+00, 9.287885e+00, 3.458050e-01, 5.616310e-01, 3.436800e+00, 3.498174e+00, 7.280371e+00, 7.562798e+00, 3.932654e+00, 5.505205e+00, 6.547320e-01, 1.499930e+00, 7.433846e+00, 3.173025e+00, 3.402000e-03, 5.562020e-01, 6.764350e-01, 7.671570e-01, 3.380080e-01, 3.988670e-01, 4.167500e-01, 1.170431e+00, 1.315025e+00, 3.612000e-03, 1.365308e+00, 1.818938e+00, 1.222941e+00, 1.896200e-02, 1.032000e-03, 1.436024e+00, 1.420145e+00]) - earthsearch:boa_offset_applied(time)boolTrue True False ... False False
array([ True, True, False, True, False, True, False, True, False, False, True, False, True, False, True, False, True, False, False, True, False, True, False, False, True, False, False, True, False, False, False, False, False, True, False, False, True, False, True, False, True, False, True, False, True, False, True, False, True, False, False, False, True, False, False, False, False, True, False, False, False, True, True, False, False]) - s2:medium_proba_clouds_percentage(time)float640.3084 2.001 4.83 ... 0.7785 6.036
array([3.0839200e-01, 2.0012220e+00, 4.8300210e+00, 1.1400400e-01, 7.8954940e+00, 5.4519200e-01, 8.7230500e-01, 7.8470000e-03, 3.4971970e+00, 6.4493430e+00, 3.2733000e-02, 6.7670320e+00, 1.0240000e-02, 6.0864860e+00, 7.0511600e-01, 2.6245750e+00, 1.1135000e-02, 1.1267800e+00, 4.5657100e-01, 6.3280500e-01, 1.2255834e+01, 2.1078810e+00, 2.8801950e+00, 1.7762980e+00, 1.1998870e+00, 2.8858000e+00, 1.4034870e+00, 2.2938500e+00, 2.1778990e+00, 3.4028490e+00, 3.4398040e+00, 1.0365550e+00, 2.0237860e+00, 5.4056610e+00, 1.7999580e+00, 2.4495680e+00, 8.4237900e-01, 1.2673150e+00, 3.3312150e+00, 1.9414730e+00, 5.5801570e+00, 2.3358590e+00, 4.6721980e+00, 3.7431200e+00, 7.7943700e-01, 2.7029620e+00, 5.7600820e+00, 6.6311990e+00, 4.4880000e-03, 6.2181100e-01, 4.2853410e+00, 9.1303400e-01, 3.8397000e-01, 9.6128100e-01, 1.1558286e+01, 3.2200780e+00, 4.9589600e-01, 7.2393000e-02, 2.5707010e+00, 2.1360510e+00, 4.1729000e+00, 1.2638000e-02, 1.4300000e-03, 7.7850800e-01, 6.0363960e+00]) - s2:vegetation_percentage(time)object1.003821 2.674679 ... 11.594474
array([1.003821, 2.674679, 2.938587, 6.434379, 7.419121, 3.186883, 3.323415, 4.962473, 5.664953, 6.822482, 6.334234, 12.347518, 11.537604, 11.502437, 10.984971, 11.278326, 16.755292, 17.688406, 20.000146, 24.586424, 26.839373, 29.225379, 31.2345, 45.217824, 40.248781, 40.689981, 64.543813, 63.144523, 67.6287, 68.127602, 64.335382, 76.903182, 69.595343, 65.299129, 68.121457, 57.554764, 59.904116, 60.774791, 56.98514, 58.596641, 55.844522, 58.502585, 46.137369, 47.877368, 62.104517, 63.009542, 47.673592, 58.927053, 59.844887, 61.083966, 47.538799, 56.53919, 54.613954, 53.221428, 25.320077, 21.666245, 27.581957, 2.711944, 3.611166, 5.17522, 10.205209, 9.482228, 10.255733, 11.158184, 11.594474], dtype=object) - s2:degraded_msi_data_percentage(time)object0.0223 0.025 0 ... 0.0039 0 0
array([0.0223, 0.025, 0, 0.0109, 0, 0.0224, 0, 0.0036, 0, 0, 0.0248, 0, 0.0231, 0, 0.0253, 0, 0.0051, 0, 0, 0.0362, 0, 0.0063, 0, 0, 0.0112, 0, 0, 0.0258, 0, 0, 0, 0, 0, 0.0059, 0, 0, 0.0108, 0, 0.0228, 0, 0.0039, 0, 0.0231, 0, 0.0037, 0, 0.0255, 0, 0.0231, 0, 0, 0, 0.0248, 0, 0, 0, 0, 0.0212, 0, 0, 0, 0.0223, 0.0039, 0, 0], dtype=object) - earthsearch:payload_id(time)<U74'roda-sentinel2/workflow-sentine...
array(['roda-sentinel2/workflow-sentinel2-to-stac/5a93f0464cc0edb8bdbbb2e7a0d2012c', 'roda-sentinel2/workflow-sentinel2-to-stac/563d331b120fee62a7f57075ebf1e004', 'roda-sentinel2/workflow-sentinel2-to-stac/598a8dec1a4123515f6b43db8c007b59', 'roda-sentinel2/workflow-sentinel2-to-stac/ff90e0b59a441d85c80dbe6fe8328957', 'roda-sentinel2/workflow-sentinel2-to-stac/98e1e06f2b2cb73eba246847a0d9b450', 'roda-sentinel2/workflow-sentinel2-to-stac/5f6f0871feb3962cd329be439c10181d', 'roda-sentinel2/workflow-sentinel2-to-stac/b950235c00b037b0f2c2148ecf0b73c7', 'roda-sentinel2/workflow-sentinel2-to-stac/4307d1487cea4f5ea795cdeea4b0fe27', 'roda-sentinel2/workflow-sentinel2-to-stac/0dd09c964b00a39216d78790364a8608', 'roda-sentinel2/workflow-sentinel2-to-stac/7ccce16aef92f0603f08f81d50f614e2', 'roda-sentinel2/workflow-sentinel2-to-stac/b39506708ba27f0f1a2b202a45bc81c0', 'roda-sentinel2/workflow-sentinel2-to-stac/21a009e0f4c71ab77557d121b4cde709', 'roda-sentinel2/workflow-sentinel2-to-stac/1d2fa679bc2794443c175233cf2e782b', 'roda-sentinel2/workflow-sentinel2-to-stac/e76bf62e0487c87d6331bce1274d3761', 'roda-sentinel2/workflow-sentinel2-to-stac/80cead34f3d3c48dcff95581129900db', 'roda-sentinel2/workflow-sentinel2-to-stac/97b157af4bdd0cc477a46d0d4ea5a860', 'roda-sentinel2/workflow-sentinel2-to-stac/811b5b0e61369b4b9b25c806a92e38e0', 'roda-sentinel2/workflow-sentinel2-to-stac/f45fdf5598dede3a3cef5a9cdb461956', 'roda-sentinel2/workflow-sentinel2-to-stac/43fc5027ff60449d117dce677c8e8135', 'roda-sentinel2/workflow-sentinel2-to-stac/4e8597d802361fd111afa86628f46741', ... 'roda-sentinel2/workflow-sentinel2-to-stac/14fde5b1530a5a0f03635435b4f9448d', 'roda-sentinel2/workflow-sentinel2-to-stac/c6f1bedd4fd9719da92f05a68da25171', 'roda-sentinel2/workflow-sentinel2-to-stac/0eb2997c603c34848e43e909afdbbd1a', 'roda-sentinel2/workflow-sentinel2-to-stac/e8f2a7d7b54105b01a394c0e85008e67', 'roda-sentinel2/workflow-sentinel2-to-stac/409bbd2ef9e61f4dec621304c7623221', 'roda-sentinel2/workflow-sentinel2-to-stac/b0bbdcb5a67e9ff1a7aa9b53a80cc8d6', 'roda-sentinel2/workflow-sentinel2-to-stac/ede35147739fd520ee13448b61b3e45c', 'roda-sentinel2/workflow-sentinel2-to-stac/1a3184f69dd0cadcfbd0df48f1392b9f', 'roda-sentinel2/workflow-sentinel2-to-stac/9c05d77aa1eff3eb9f854a50c035550b', 'roda-sentinel2/workflow-sentinel2-to-stac/7ab374294204a80b9bef95d5583df9a6', 'roda-sentinel2/workflow-sentinel2-to-stac/3153efafe0c48eccb67f4657900bf6b9', 'roda-sentinel2/workflow-sentinel2-to-stac/93568ff358f842879ea566c2e766d45d', 'roda-sentinel2/workflow-sentinel2-to-stac/71aba90ac7b3a173c126c68065ea5637', 'roda-sentinel2/workflow-sentinel2-to-stac/8366e35ded0e5bd249b6580f59a52c49', 'roda-sentinel2/workflow-sentinel2-to-stac/ad99eb1e478397b6092c8f33bfb2155a', 'roda-sentinel2/workflow-sentinel2-to-stac/62fdb0869502d7847df53e10633177e6', 'roda-sentinel2/workflow-sentinel2-to-stac/a60d1d53197ebe894647143a010a1bb6', 'roda-sentinel2/workflow-sentinel2-to-stac/313a6f02840be6a2663a0e3230e357b2', 'roda-sentinel2/workflow-sentinel2-to-stac/c821731deb57cc090c7762e61fba8553'], dtype='<U74') - gsd()int6410
array(10)
- proj:shape()object{10980}
array({10980}, dtype=object) - proj:transform()object{0, 600000, 10, 5200020, -10}
array({0, 600000, 10, 5200020, -10}, dtype=object) - epsg()int6432632
array(32632)
- timePandasIndex
PandasIndex(DatetimeIndex(['2021-01-11 10:27:54.911000', '2021-02-15 10:27:54.551000', '2021-02-15 10:27:54.552000', '2021-02-17 10:17:57.062000', '2021-02-17 10:17:57.062000', '2021-02-20 10:27:53.334000', '2021-02-20 10:27:53.335000', '2021-02-22 10:17:58.873000', '2021-02-22 10:17:58.873000', '2021-02-25 10:27:55.240000', '2021-02-25 10:27:55.241000', '2021-03-02 10:27:54.438000', '2021-03-02 10:27:54.439000', '2021-03-07 10:27:55.298000', '2021-03-07 10:27:55.300000', '2021-03-22 10:27:54.580000', '2021-03-24 10:17:57.711000', '2021-03-24 10:17:57.712000', '2021-04-01 10:27:53.659000', '2021-04-08 10:17:56.404000', '2021-04-08 10:17:56.404000', '2021-04-23 10:17:55.224000', '2021-04-23 10:17:55.227000', '2021-05-26 10:27:56.205000', '2021-05-28 10:17:59.882000', '2021-05-28 10:17:59.884000', '2021-06-15 10:27:55.829000', '2021-06-15 10:27:55.830000', '2021-06-27 10:18:00.078000', '2021-07-12 10:18:02.331000', '2021-07-20 10:27:57.564000', '2021-08-14 10:27:58.017000', '2021-08-26 10:17:57.394000', '2021-09-10 10:17:59.484000', '2021-09-10 10:17:59.484000', '2021-09-18 10:27:51.968000', '2021-09-25 10:17:57.577000', '2021-09-25 10:17:57.578000', '2021-09-28 10:27:54.449000', '2021-09-28 10:27:54.449000', '2021-09-30 10:18:03.604000', '2021-09-30 10:18:03.604000', '2021-10-08 10:27:56.160000', '2021-10-08 10:27:56.160000', '2021-10-10 10:18:04.559000', '2021-10-10 10:18:04.560000', '2021-10-13 10:28:00.872000', '2021-10-15 10:18:00.588000', '2021-10-18 10:27:56.971000', '2021-10-18 10:27:56.973000', '2021-10-20 10:18:04.648000', '2021-10-23 10:28:00.726000', '2021-10-23 10:28:00.728000', '2021-10-28 10:27:56.929000', '2021-11-07 10:27:55.834000', '2021-11-12 10:27:57.394000', '2021-11-24 10:17:55.331000', '2021-12-12 10:27:49.590000', '2021-12-12 10:27:49.591000', '2021-12-12 10:27:56.297000', '2021-12-17 10:27:49.270000', '2021-12-17 10:27:49.271000', '2021-12-19 10:17:59.960000', '2021-12-19 10:17:59.962000', '2021-12-22 10:27:56.749000'], dtype='datetime64[ns]', name='time', freq=None))
Remember that calling .load() on an xarray + dask based object will load into memory the data.
In this case it will trigger the download of the data from the STAC Catalog and the processing defined as openEO process graph, computing the NDVI time series.
Visualize the NDVI time series:
ndvi_spatial_xr.where(ndvi_spatial_xr<1).plot.scatter()
<matplotlib.collections.PathCollection at 0x7fc396776690>