Apply Operator with Pangeo#
The apply operator employ a process on the datacube that calculates new pixel values for each pixel, based on n other pixels.
Let’s start again with the same sample data from the Sentinel-2 STAC Collection, applying the filters directly in the stackstac.stack call, to reduce the amount of data.
# STAC Catalogue Libraries
import pystac_client
import stackstac
# West, South, East, North
spatial_extent = [11.259613, 46.461019, 11.406212, 46.522237]
temporal_extent = ['2022-07-10T00:00:00Z','2022-07-13T00:00:00Z']
bands = ["red","green","blue"]
URL = "https://earth-search.aws.element84.com/v1"
catalog = pystac_client.Client.open(URL)
s2_items = catalog.search(
bbox=spatial_extent,
datetime=temporal_extent,
query=["eo:cloud_cover<50"],
collections=["sentinel-2-l2a"]
).item_collection()
s2_cube = stackstac.stack(s2_items,
bounds_latlon=spatial_extent,
assets=bands
)
s2_cube
/srv/conda/envs/notebook/lib/python3.11/site-packages/stackstac/prepare.py:408: UserWarning: The argument 'infer_datetime_format' is deprecated and will be removed in a future version. A strict version of it is now the default, see https://pandas.pydata.org/pdeps/0004-consistent-to-datetime-parsing.html. You can safely remove this argument.
times = pd.to_datetime(
<xarray.DataArray 'stackstac-0747b87a1b91037120a189227e8f5908' (time: 1,
band: 3,
y: 714, x: 1146)>
dask.array<fetch_raster_window, shape=(1, 3, 714, 1146), dtype=float64, chunksize=(1, 1, 714, 1024), chunktype=numpy.ndarray>
Coordinates: (12/54)
* time (time) datetime64[ns] 2022-07-12...
id (time) <U24 'S2B_32TPS_20220712_...
* band (band) <U5 'red' 'green' 'blue'
* x (x) float64 6.733e+05 ... 6.848e+05
* y (y) float64 5.155e+06 ... 5.148e+06
view:sun_azimuth float64 146.9
... ...
proj:shape object {10980}
gsd int64 10
common_name (band) <U5 'red' 'green' 'blue'
center_wavelength (band) float64 0.665 0.56 0.49
full_width_half_max (band) float64 0.038 0.045 0.098
epsg int64 32632
Attributes:
spec: RasterSpec(epsg=32632, bounds=(673310.0, 5147750.0, 684770.0...
crs: epsg:32632
transform: | 10.00, 0.00, 673310.00|\n| 0.00,-10.00, 5154890.00|\n| 0.0...
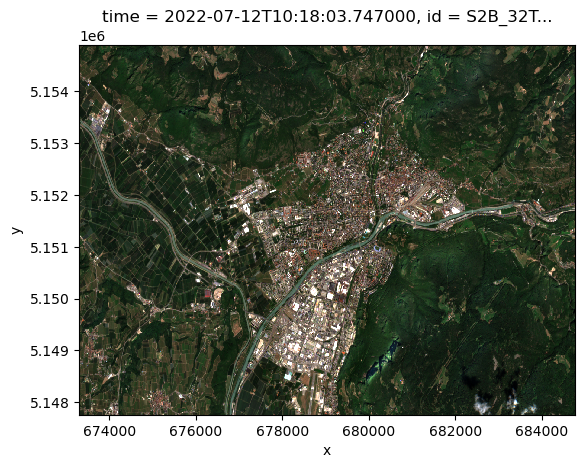
resolution: 10.0Visualize the RGB bands of our sample dataset:
s2_cube.isel(time=0).plot.imshow()
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
<matplotlib.image.AxesImage at 0x7ff48857fd90>
Apply an arithmetic formula#
We would like to improve the previous visualization, rescaling all the pixels between 0 and 1.
We can use apply in combination with other math processes.
s2_cube
<xarray.DataArray 'stackstac-0747b87a1b91037120a189227e8f5908' (time: 1,
band: 3,
y: 714, x: 1146)>
dask.array<fetch_raster_window, shape=(1, 3, 714, 1146), dtype=float64, chunksize=(1, 1, 714, 1024), chunktype=numpy.ndarray>
Coordinates: (12/54)
* time (time) datetime64[ns] 2022-07-12...
id (time) <U24 'S2B_32TPS_20220712_...
* band (band) <U5 'red' 'green' 'blue'
* x (x) float64 6.733e+05 ... 6.848e+05
* y (y) float64 5.155e+06 ... 5.148e+06
view:sun_azimuth float64 146.9
... ...
proj:shape object {10980}
gsd int64 10
common_name (band) <U5 'red' 'green' 'blue'
center_wavelength (band) float64 0.665 0.56 0.49
full_width_half_max (band) float64 0.038 0.045 0.098
epsg int64 32632
Attributes:
spec: RasterSpec(epsg=32632, bounds=(673310.0, 5147750.0, 684770.0...
crs: epsg:32632
transform: | 10.00, 0.00, 673310.00|\n| 0.00,-10.00, 5154890.00|\n| 0.0...
resolution: 10.0import xarray as xr
input_min = -0.1
input_max = 0.2
output_min = 0
output_max = 1
def rescale(x, input_min, input_max, output_min, output_max):
y = xr.where(x>=input_min, x, input_min)
y = xr.where(y<=input_max, y, input_max)
return ((y - input_min) / (input_max - input_min)) * (output_max - output_min) + output_min
scaled_data = rescale(s2_cube, input_min, input_max, output_min, output_max)
input_min = -0.1
input_max = 0.2
output_min = 0
output_max = 1
def rescale(x, input_min, input_max, output_min, output_max):
return ((x.where(x>=input_min, input_min).where(x<=input_max, input_max) - input_min) / (input_max - input_min)) * (output_max - output_min) + output_min
scaled_data = rescale(s2_cube, input_min, input_max, output_min, output_max)
scaled_data.load()
<xarray.DataArray 'stackstac-0747b87a1b91037120a189227e8f5908' (time: 1,
band: 3,
y: 714, x: 1146)>
array([[[[0.218 , 0.20266667, 0.20733333, ..., 0.138 ,
0.095 , 0.11733333],
[0.252 , 0.24733333, 0.21933333, ..., 0.08866667,
0.07866667, 0.07566667],
[0.206 , 0.16066667, 0.19733333, ..., 0.07 ,
0.08 , 0.08066667],
...,
[0.23933333, 0.29166667, 0.12533333, ..., 0.072 ,
0.07833333, 0.072 ],
[0.196 , 0.30666667, 0.16666667, ..., 0.07733333,
0.069 , 0.05133333],
[0.178 , 0.28933333, 0.22066667, ..., 0.06766667,
0.05966667, 0.058 ]],
[[0.21566667, 0.19466667, 0.21133333, ..., 0.152 ,
0.14466667, 0.15633333],
[0.226 , 0.21133333, 0.23 , ..., 0.13233333,
0.137 , 0.12533333],
[0.21466667, 0.17 , 0.184 , ..., 0.13266667,
0.13966667, 0.121 ],
...
[0.296 , 0.29533333, 0.15933333, ..., 0.146 ,
0.16166667, 0.13133333],
[0.24 , 0.326 , 0.19666667, ..., 0.15633333,
0.14 , 0.117 ],
[0.24133333, 0.33133333, 0.21666667, ..., 0.14866667,
0.139 , 0.13066667]],
[[0.162 , 0.13633333, 0.15033333, ..., 0.08933333,
0.08566667, 0.08533333],
[0.16166667, 0.171 , 0.18 , ..., 0.06366667,
0.06866667, 0.07333333],
[0.14033333, 0.117 , 0.14733333, ..., 0.076 ,
0.08 , 0.074 ],
...,
[0.19666667, 0.24166667, 0.10166667, ..., 0.073 ,
0.08366667, 0.06866667],
[0.12966667, 0.26733333, 0.11733333, ..., 0.07866667,
0.08233333, 0.07333333],
[0.2 , 0.21466667, 0.183 , ..., 0.08666667,
0.07966667, 0.07533333]]]])
Coordinates: (12/54)
* time (time) datetime64[ns] 2022-07-12...
id (time) <U24 'S2B_32TPS_20220712_...
* band (band) <U5 'red' 'green' 'blue'
* x (x) float64 6.733e+05 ... 6.848e+05
* y (y) float64 5.155e+06 ... 5.148e+06
view:sun_azimuth float64 146.9
... ...
proj:shape object {10980}
gsd int64 10
common_name (band) <U5 'red' 'green' 'blue'
center_wavelength (band) float64 0.665 0.56 0.49
full_width_half_max (band) float64 0.038 0.045 0.098
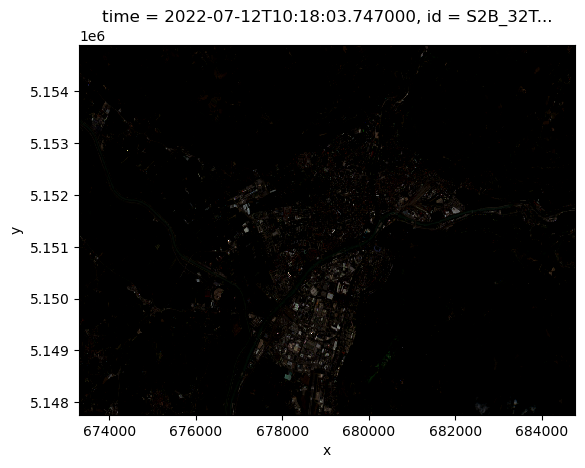
epsg int64 32632Visualize the result and see how apply scaled the data resulting in a more meaningful visualization:
scaled_data.isel(time=0).plot.imshow()
<matplotlib.image.AxesImage at 0x7ff48857f910>