pangeo_CMIP6_masked_and_weighted_average
Using masks and computing weighted average
This example is based from xarray example http://xarray.pydata.org/en/stable/examples/area_weighted_temperature.html
Import python packages
pip install nc-time-axis
Requirement already satisfied: nc-time-axis in /usr/share/miniconda/envs/repo/lib/python3.10/site-packages (1.4.1)
Requirement already satisfied: matplotlib in /usr/share/miniconda/envs/repo/lib/python3.10/site-packages (from nc-time-axis) (3.5.2)
Requirement already satisfied: cftime>=1.5 in /usr/share/miniconda/envs/repo/lib/python3.10/site-packages (from nc-time-axis) (1.6.0)
Requirement already satisfied: numpy in /usr/share/miniconda/envs/repo/lib/python3.10/site-packages (from nc-time-axis) (1.22.4)
Requirement already satisfied: pyparsing>=2.2.1 in /usr/share/miniconda/envs/repo/lib/python3.10/site-packages (from matplotlib->nc-time-axis) (3.0.9)
Requirement already satisfied: python-dateutil>=2.7 in /usr/share/miniconda/envs/repo/lib/python3.10/site-packages (from matplotlib->nc-time-axis) (2.8.2)
Requirement already satisfied: pillow>=6.2.0 in /usr/share/miniconda/envs/repo/lib/python3.10/site-packages (from matplotlib->nc-time-axis) (9.1.1)
Requirement already satisfied: fonttools>=4.22.0 in /usr/share/miniconda/envs/repo/lib/python3.10/site-packages (from matplotlib->nc-time-axis) (4.33.3)
Requirement already satisfied: packaging>=20.0 in /usr/share/miniconda/envs/repo/lib/python3.10/site-packages (from matplotlib->nc-time-axis) (21.3)
Requirement already satisfied: kiwisolver>=1.0.1 in /usr/share/miniconda/envs/repo/lib/python3.10/site-packages (from matplotlib->nc-time-axis) (1.4.2)
Requirement already satisfied: cycler>=0.10 in /usr/share/miniconda/envs/repo/lib/python3.10/site-packages (from matplotlib->nc-time-axis) (0.11.0)
Requirement already satisfied: six>=1.5 in /usr/share/miniconda/envs/repo/lib/python3.10/site-packages (from python-dateutil>=2.7->matplotlib->nc-time-axis) (1.16.0)
Note: you may need to restart the kernel to use updated packages.
import xarray as xr
xr.set_options(display_style='html')
import intake
import cftime
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import numpy as np
%matplotlib inline
cat_url = "https://storage.googleapis.com/cmip6/pangeo-cmip6.json"
col = intake.open_esm_datastore(cat_url)
col
pangeo-cmip6 catalog with 7686 dataset(s) from 514961 asset(s):
| unique | |
|---|---|
| activity_id | 18 |
| institution_id | 36 |
| source_id | 88 |
| experiment_id | 170 |
| member_id | 657 |
| table_id | 37 |
| variable_id | 700 |
| grid_label | 10 |
| zstore | 514961 |
| dcpp_init_year | 60 |
| version | 737 |
Search data
cat = col.search(source_id=['NorESM2-LM'], experiment_id=['historical'], table_id=['Amon'], variable_id=['tas'], member_id=['r1i1p1f1'])
cat.df
| activity_id | institution_id | source_id | experiment_id | member_id | table_id | variable_id | grid_label | zstore | dcpp_init_year | version | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | CMIP | NCC | NorESM2-LM | historical | r1i1p1f1 | Amon | tas | gn | gs://cmip6/CMIP6/CMIP/NCC/NorESM2-LM/historica... | NaN | 20190815 |
Create dictionary from the list of datasets we found
- This step may take several minutes so be patient!
dset_dict = cat.to_dataset_dict(zarr_kwargs={'use_cftime':True})
--> The keys in the returned dictionary of datasets are constructed as follows:
'activity_id.institution_id.source_id.experiment_id.table_id.grid_label'
100.00% [1/1 00:00<00:00]
list(dset_dict.keys())
['CMIP.NCC.NorESM2-LM.historical.Amon.gn']
dset = dset_dict[list(dset_dict.keys())[0]]
dset
<xarray.Dataset>
Dimensions: (lat: 96, bnds: 2, lon: 144, member_id: 1, time: 1980)
Coordinates:
height float64 ...
* lat (lat) float64 -90.0 -88.11 -86.21 -84.32 ... 86.21 88.11 90.0
lat_bnds (lat, bnds) float64 dask.array<chunksize=(96, 2), meta=np.ndarray>
* lon (lon) float64 0.0 2.5 5.0 7.5 10.0 ... 350.0 352.5 355.0 357.5
lon_bnds (lon, bnds) float64 dask.array<chunksize=(144, 2), meta=np.ndarray>
* time (time) object 1850-01-16 12:00:00 ... 2014-12-16 12:00:00
time_bnds (time, bnds) object dask.array<chunksize=(1980, 2), meta=np.ndarray>
* member_id (member_id) <U8 'r1i1p1f1'
Dimensions without coordinates: bnds
Data variables:
tas (member_id, time, lat, lon) float32 dask.array<chunksize=(1, 990, 96, 144), meta=np.ndarray>
Attributes: (12/54)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: Hybrid-restart from year 1600-01-01 of piControl
branch_time: 0.0
branch_time_in_child: 0.0
branch_time_in_parent: 430335.0
... ...
variable_id: tas
variant_label: r1i1p1f1
netcdf_tracking_ids: hdl:21.14100/2486cf87-033c-4848-ab3e-e828c3b7c...
version_id: v20190815
intake_esm_varname: ['tas']
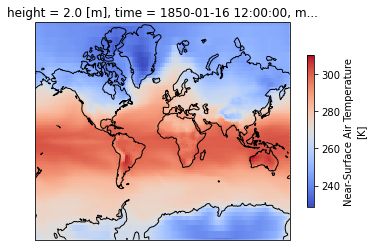
intake_esm_dataset_key: CMIP.NCC.NorESM2-LM.historical.Amon.gnPlot the first timestep
projection = ccrs.Mercator(central_longitude=-10)
f, ax = plt.subplots(subplot_kw=dict(projection=projection))
dset['tas'].isel(time=0).plot(transform=ccrs.PlateCarree(), cbar_kwargs=dict(shrink=0.7), cmap='coolwarm')
ax.coastlines()
<cartopy.mpl.feature_artist.FeatureArtist at 0x7f1212118670>
Compute weighted mean
- Creating weights: for a rectangular grid the cosine of the latitude is proportional to the grid cell area.
- Compute weighted mean values
def computeWeightedMean(ds):
# Compute weights based on the xarray you pass
weights = np.cos(np.deg2rad(ds.lat))
weights.name = "weights"
# Compute weighted mean
air_weighted = ds.weighted(weights)
weighted_mean = air_weighted.mean(("lon", "lat"))
return weighted_mean
Compute weighted average over the entire globe
weighted_mean = computeWeightedMean(dset)
Comparison with unweighted mean
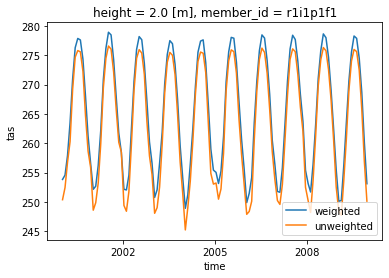
- We select a time range
- Note how the weighted mean temperature is higher than the unweighted.
weighted_mean['tas'].sel(time=slice('2000-01-01', '2010-01-01')).plot(label="weighted")
dset['tas'].sel(time=slice('2000-01-01', '2010-01-01')).mean(("lon", "lat")).plot(label="unweighted")
plt.legend()
<matplotlib.legend.Legend at 0x7f12138a66b0>
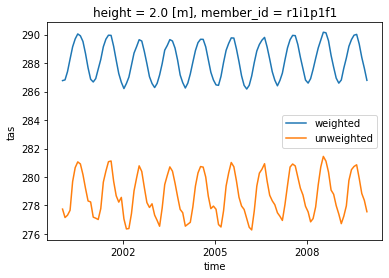
Compute Weigted arctic average
Let’s try to also take only the data above 60$^\circ$
weighted_mean = computeWeightedMean(dset.where(dset['lat']>60.))
weighted_mean['tas'].sel(time=slice('2000-01-01', '2010-01-01')).plot(label="weighted")
dset['tas'].where(dset['lat']>60.).sel(time=slice('2000-01-01', '2010-01-01')).mean(("lon", "lat")).plot(label="unweighted")
plt.legend()
<matplotlib.legend.Legend at 0x7f1211b59480>